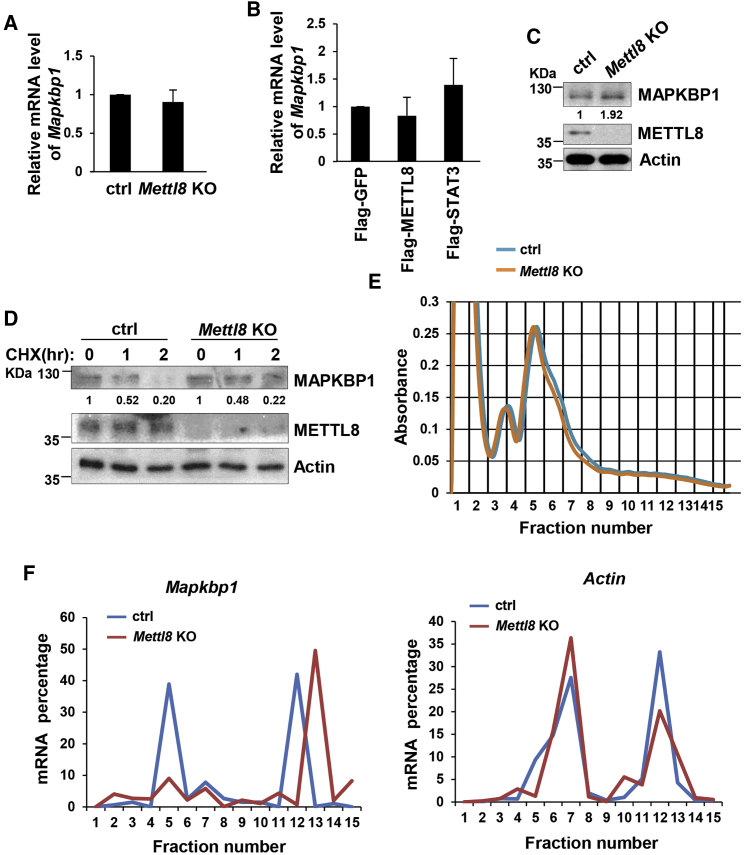
Figure 5.
Mettl8 Deficiency Enhances Mapkbp1 mRNA Translation
(A) Total RNAs were extracted from Mettl8 KO and control E14 cells followed by real-time PCR analysis. Data are shown as the mean ± SD from three independent experiments.
(B) HEK293T cells were transfected by indicated plasmids. After 48 hr total RNAs were extracted followed by real-time PCR analysis. Data are shown as the mean ± SD from three independent experiments.
(C) Cell lysates of Mettl8 KO and control E14 cells were analyzed by western blot. The value of each band was calculated from three independent replicates and indicates the relative expression level after normalizing to the loading control actin.
(D) Mettl8 KO and control E14 cells were treated with CHX for the indicated times and cell lysates were analyzed by western blot. The value of each band was calculated from three independent replicates and indicates the relative expression level after normalizing to the loading control actin.
(E) Global polysome profiles of Mettl8 KO and control E14 cells were analyzed by sucrose gradient sedimentation.
(F) Each fraction from sucrose gradient sedimentation was collected and total RNA was extracted. Levels of Mapkbp1 mRNA were analyzed by quantitative real-time PCR.