Figure 1.
Analysis of Induced Pluripotent Stem Cell-Derived Endothelial Cells Using Convolutional Neural Networks
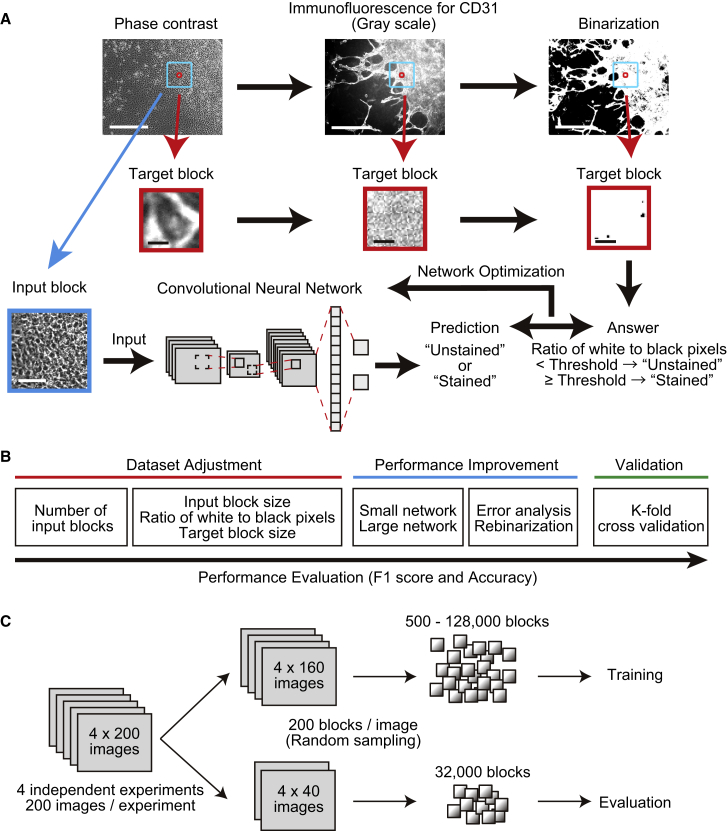
(A) Training protocol. Input blocks were extracted from phase-contrast images and predicted by networks to be unstained (0) or stained (1) for CD31. Target blocks containing single cells were extracted from immunofluorescent images of the same field, binarized based on CD31 staining, and classified as stained or unstained based on the ratio of white pixels to black. Network weights were then automatically and iteratively adjusted to maximize agreement between predicted and observed classification. Scale bars, 400 μm (upper panels), 5 μm (middle panels), and 80 μm (bottom panels).
(B) Optimization of experimental parameters to maximize F1 score and accuracy.
(C) Two hundred images each were obtained from four independent experiments. Images were randomized at 80:20 ratio into training and evaluation sets, and 200 blocks were randomly extracted from each image.