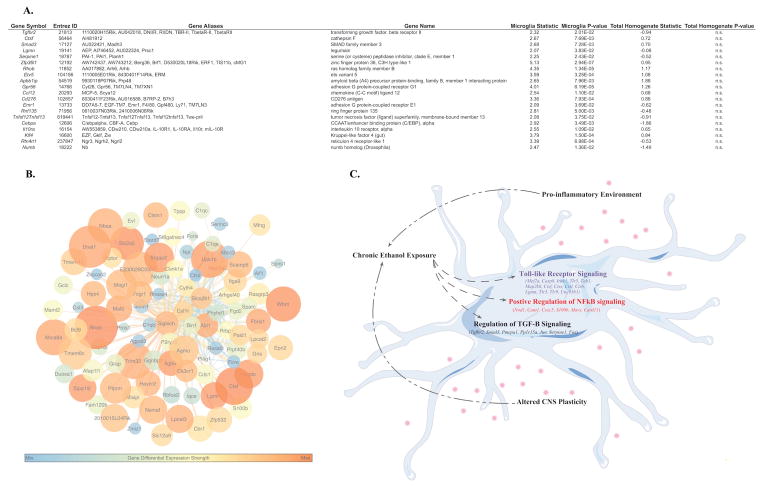
Figure 5.
A. Table of differentially expressed genes in MGM3. Each listing contains the gene symbol., Entrez ID, Gene Aliases and name, the T-statistic and p-value for each gene in microglia and the total homogenate. All genes shown in the table were differentially expressed with ethanol in microglia but not in the total homogenate. B. Connectivity map containing the top 250 connections in MGM3. Warmer colored nodes have smaller p-values for differential expression with alcohol. Larger nodes have larger fold changes with alcohol. Larger connecting lines indicate higher topological overlap measure (TOM), or connectivity between 2 genes. Warmer colored lines indicate higher edge betweenness centrality, or importance of the connection in the network. Siglech is highly connected to many genes in MGM3. C. Hypothesis about effects of chronic ethanol on microglia. Chronic ethanol exposure leads to altered expression of genes involved in Toll-like Receptor Signaling, NF-κB signaling, and TGF-β signaling. Together, these alterations in immune signaling can result in a pro-inflammatory environment and altered CNS plasticity, which can drive further consumption.