Figure 4. Reference perturbagen classes for CMap discovery.
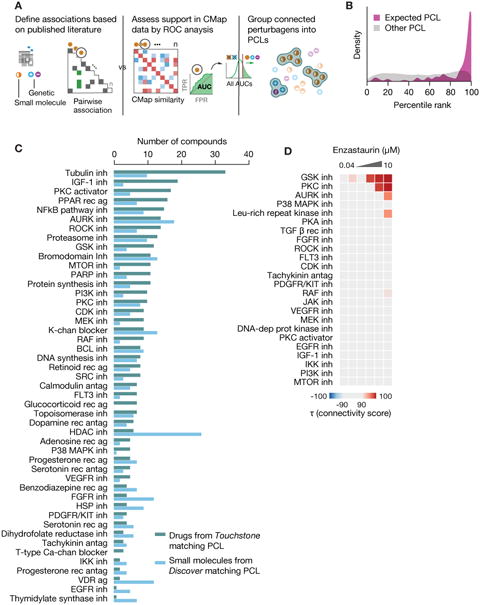
A. Process for defining Perturbagen Classes (PCLs). Left: Annotations gathered from literature sources to construct pairwise association matrix between perturbagens based on shared descriptors such as MoA, target gene and pathway membership. Middle: Each perturbagen is subject to ROC analysis to determine whether it recovers expected connections. Right: Remaining members are grouped based on shared annotations and assessed for intra-group connectivity of CMap signatures. Groups sufficiently interconnected are retained as PCLs.
B. PCL validation. 137 compounds with known activities corresponding to one or more of 54 PCLs, but not used in PCL construction, were profiled across multiple cell types. Histogram shows rank of each expected PCL connection for the compounds (purple) versus the rank of all unexpected PCL connections (grey). The expected PCL distribution is significantly right-shifted (one-sided p < 2.2e-16 via two-sample KS test).
C. Using PCLs for discovery. 3,333 known drugs and 2,418 unannotated but transcriptionally active compounds were subject to PCL analysis. Count of strong and selective connections to validated PCLs byknown drugs (teal) and unannotated compounds (blue). Abbreviations: inh. inhibitor, ag. agonist, rec.receptor, antag. antagonist, and chan. channel.
D. Detecting multiple drug activities using PCLs. The PKC inhibitor enzastaurin was profiled in CMap across multiple doses. Connectivity to each established kinase inhibitor PCL is shown in the heatmap. Strong dose-responsive connections were observed to PKC and GSK3 inhibitor PCLs.
