Figure 7. Assessing impact of allelic variants and drug response in clinical trials.
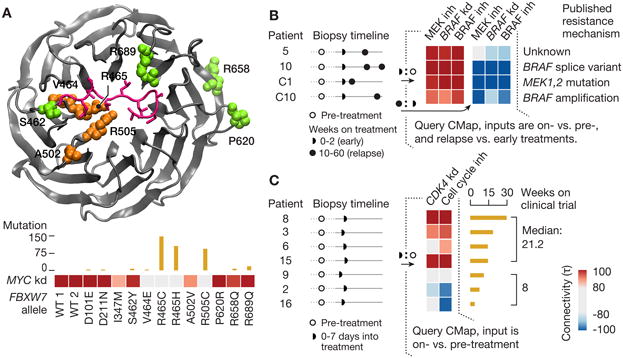
A. Predicting LoF alleles. Clinically-observed FBXW7 alleles were overexpressed and L1000 profiles obtained. Protein structure shows residues in question. Wild-type FBXW7 connects strongly to MYC shRNA, which is a known target (heat map). Mutations at residues adjacent to the substrate recognition site lose the MYC connection. τ values are summarized across multiple cell types. Bar plot above heat map indicates incidence of each mutation in COSMIC database.
B. Interpreting drug resistance. Transcriptional profiles of pre-treatment, early on-treatment, and relapse tumor biopsies obtained from clinical trials of BRAF and MEK inhibitors. Queries from on-treatment versus pre-treatment biopsies exhibited connectivity to pharmacologic inhibition of BRAF or MEK as well as BRAF shRNA in A375 cells, reflecting target engagement in vivo (left 3 columns in heat map). MAP kinase signaling was re-activated, as indicated by a strong negative connection to the same CMap signature in the subset of relapse biopsies with known MAP kinase pathway-related resistance mutations (right 3 columns of heat map).
C. Predicting therapeutic efficacy. Transcriptional profiles of pre-treatment and on-treatment biopsies from clinical trial of PHA-793887. Differential expression between the two time points yielded variable connectivity to negative regulators of cell cycle. Patients with strong positive connectivity to cell cycle inhibition signatures remained on trial for a median of 21 weeks; patients with negative connections remained on study for only 8 weeks.
