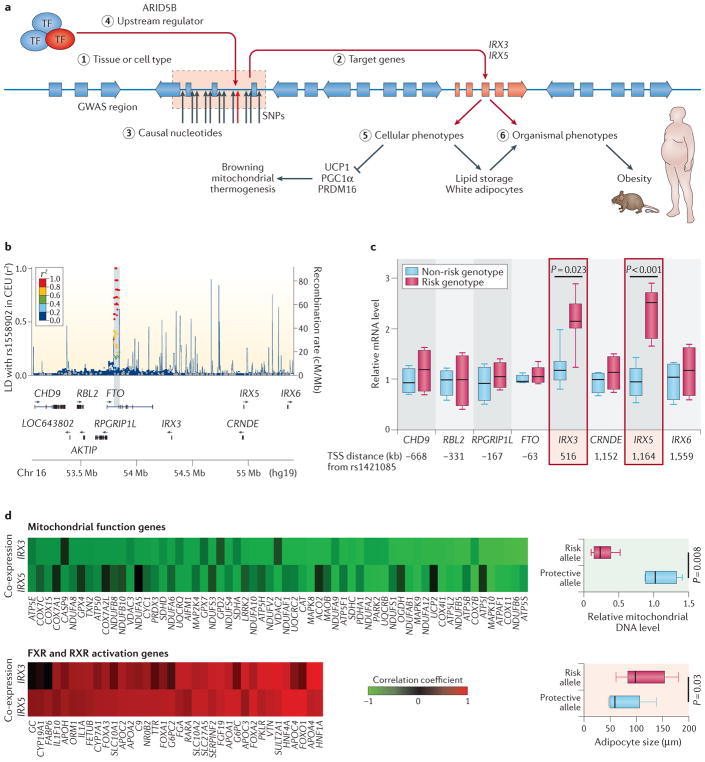
Figure 2. From genome-wide association studies to mechanism.
In a recent study, Claussnitzer and colleagues present a comprehensive approach50 to identifying a causal mechanism for an obesity-associated variant in the FTO gene. Part a shows an overview of the deciphered biological mechanisms and the numbered steps of the strategy referred to below. From the initial genome-wide association study (GWAS), the significant association of the FTO region with obesity is shown in the Manhattan plot (part b). First, the researchers established the relevant tissue or cell type (step 1) as well as the downstream target genes using regulatory genomics, including chromatin state information and chromosomal conformation (Hi-C) data. Here, they established the variant as an expression quantitative trait locus (eQTL) for the developmental genes iroquois homeobox 3 (IRX3) and IRX5 (step 2), where the risk allele shows increased expression of these genes but not others in the vicinity (part c). They demonstrate that expression of IRX3 and IRX5 is anti-correlated and correlated with genes involved in mitochondrial function and adipocyte size, respectively (part d). Next, they established the causal nucleotide variant (step 3) in an AT-rich interactive domain-containing protein 5B (ARID5B) motif (step 4) using CRISPR–Cas9 to show its molecular effects, including altered signatures of expression and phenotypic effects on the regulation of energy balance (step 5). Finally, they establish causality of the variant on an organismal level using mouse models (step 6). AKTIP, AKT interacting protein; CEU, Utah residents (CEPH) with northern and western European ancestry; CHD9, chromodomain helicase DNA binding protein 9; CRNDE, colorectal neoplasia differentially expressed; FXR, farnesoid X-activated receptor; LD, linkage disequilibrium; PGC1α, peroxisome proliferator-activated receptor-γ co-activator 1-α; PRDM16, PR domain zinc-finger protein 16; RBL2, RB transcriptional co-repressor like 2; RXR, retinoid X receptor; SNPs, single-nucleotide polymorphisms; TF, transcription factor; TSS, transcription start site; UCP1, mitochondrial brown fat uncoupling protein 1. Figure is adapted from The New England Journal of Medicine, Claussnitzer, M. et al., FTO obesity variant circuitry and adipocyte browning in humans, 373, 895–907, Copyright© (2015) Massachusetts Medical Society, REF. 50. Reprinted with permission from Massachusetts Medical Society.