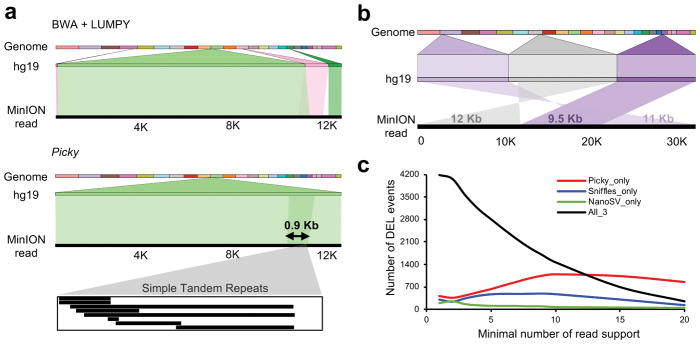
Figure 2. The sensitivity of the Picky pipeline in SV detection.
(a) Picky accurately defines a short TD. A 12.9 Kb nanopore read aligns to reference genome as two overlapping segments of 11.4 Kb and 2.4 Kb with 86% and 83% identities (e-value = 0). This TD was misclassified as a translocation by the short-read aligner BWA and SV caller LUMPY. (b) A complex SV of two translocations detected by Picky from a 32.5 Kb nanopore read. The alignments were visualized by Ribbon (https://github.com/MariaNattestad/ribbon). (c) Numbers of reference deletions supported by each versus all pipelines among different thresholds.