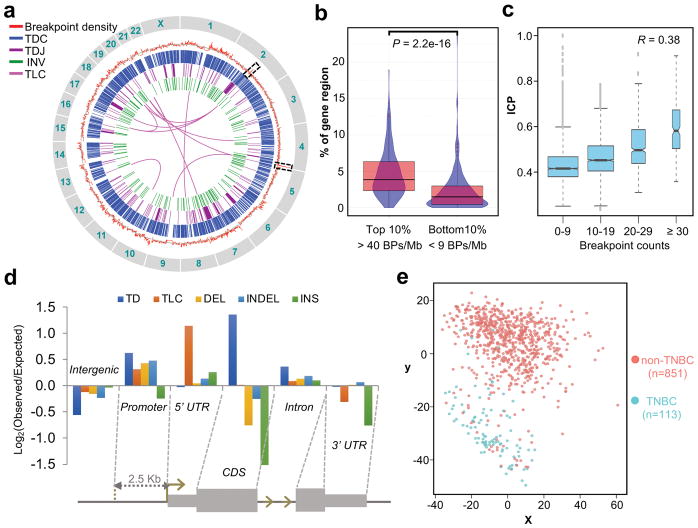
Figure 4. The analysis of genomic distribution of breakpoints and their affected genes.
(a) Genome-wide distribution of the SVs and their associated breakpoints density. From outer circle to inner circle: Red: measured breakpoint density; Grey: reference average density of 22 breakpoints/Mb; Blue: TDC; Purple: TDJ (with span size < 20 Mb); Green: INV; Magenta: TLC (with pair counts ≥ 4) at 1Mb bin size. (b) Fraction of regions with transcribed genes in regions of the top 10% (n = 250) and the bottom 10% (n = 234) breakpoint densities. BP, breakpoint. P value was calculated by Mann-Whitney test. Center line, median; boxes, first and third quartiles; whiskers, 1.5 interquartile range (IQR) from the box. (c) The trend of increasing ICP with increasing number of breakpoints. The breakpoints and ICP were calculated for every chromosome segments of 138 HindIII sites (see Online Methods). The ICP values were then grouped based on the breakpoint counts as indicated on the x-axis. 0–9, n = 2,981; 10–19, n = 2,202; 20–29, n = 622; ≥ 30, n = 197. The R value is the Pearson’s correlation coefficient on the underlying, ungrouped data. Center line, median; boxes, first and third quartiles; whiskers, 1.5 interquartile range (IQR) from the box; notch, 95% confidence interval of the median. (d) Enrichment of breakpoint from each SV class distributed along different genomic features. (e) The multidimensional scaling (MDS) plot of SV-genes in the BRCA dataset within the TCGA. Non-TNBC, n = 851; TNBC, n = 113.