Figure 6. Description of iterative integrative modeling workflow.
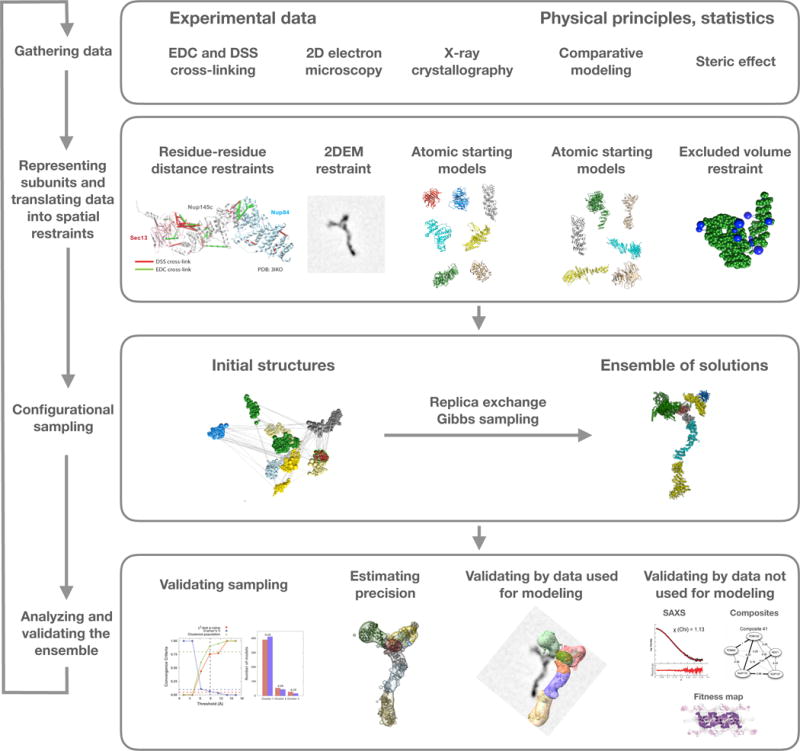
The integrative modeling workflow is illustrated by its application to structure determination of the Nup84 heptamer (Shi et al., 2014). The four stages include: (1) gathering all available experimental data and theoretical information; (2) translating this information into representations of assembly components and a scoring function for ranking alternative assembly structures; (3) sampling and scoring of structural models; and (4) analyzing and assessing the models. In this case, representations of the seven components of the Nup84 complex are based on crystallographic structures and comparative models of their domains. Component representations are coarse-grained by using spherical beads corresponding to multiple amino acid residues, to reflect the lack of information and/or to increase efficiency of sampling. The scoring function consists of spatial restraints that are obtained from CX-MS experiments and 2DEM class average images. The sampling explores both the conformations of the components and/or their configuration, searching for those assembly structures that satisfy the spatial restraints as accurately as possible. In this case, the result is an ensemble of many good-scoring models that satisfy the input data within acceptable thresholds. The sampling is then assessed for convergence, models are clustered, and evaluated by the degree to which they satisfy the data used to construct them as well as omitted data. The protocol can iterate through the four stages, until the models are judged to be satisfactory, most often based on their precision and the degree to which they satisfy the data. Finally, the models are deposited in PDB-Dev (https://pdb-dev.wwpdb.org, section 2.4).
