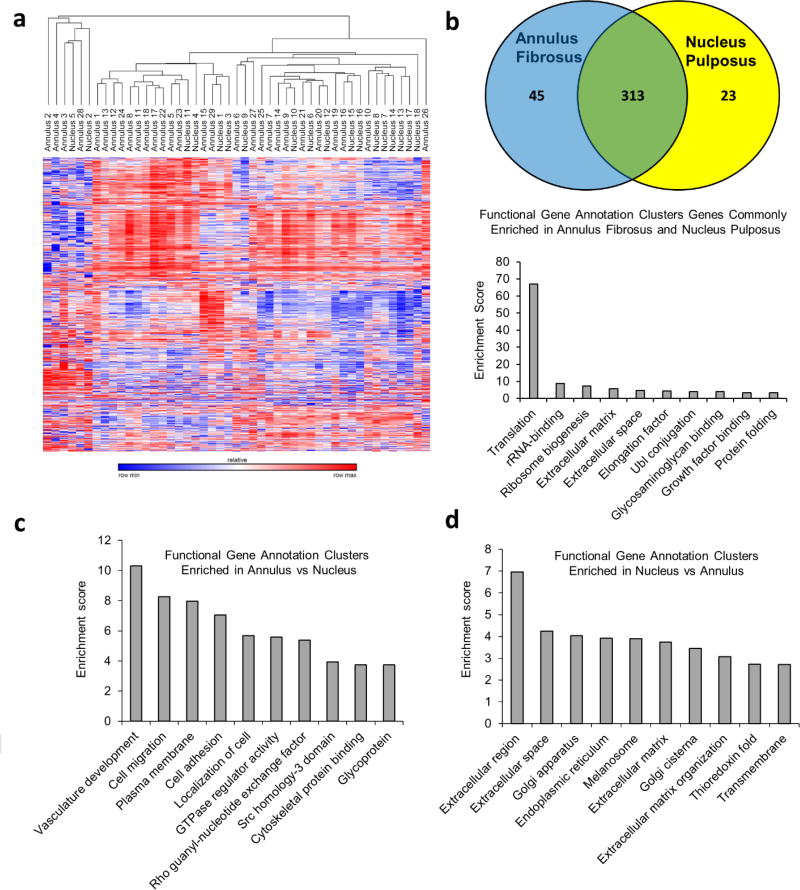
Figure 1.
(a) Unsupervised hierarchical clustering of RNA sequencing data after removal of sample outliers. In this clustering scheme there is a trend for AF and NP samples to preferentially cluster separately. These findings suggest that there are tissue specific differences contained within the transcriptome data, representing the known biological differences that exist between these two tissue types. Our unbiased approach also incorporates various factors that are not directly related to the disc phenotype such as tissue heterogeneity, blood content, and inflammation, which can drive some of the biological variation between specimens, thus precluding a perfect clustering dendogram in which AF and NP specimens cluster as completely independent groups. (b) Genes expressed > 100 RPKM in surgically isolated AF and NP tissue with equal expression levels (Fold change <1.5 between AF and NP). This analysis shows that AF and NP both share common expression of a large number of housekeeping genes as well as a small number of extracellular matrix proteins and growth factor binding associated proteins. (c) Gene ontology analysis reveals enrichment in pathways that promote cellular adhesion including genes linked to notch signaling (vasculature development, GTPase regulator activity) in AF tissue. (d) The NP shows enrichment in genes linked to extracellular matrix protein synthesis, including in genes controlling the extracellular matrix protein synthesis machinery (golgi complex and endoplasmic reticulum).