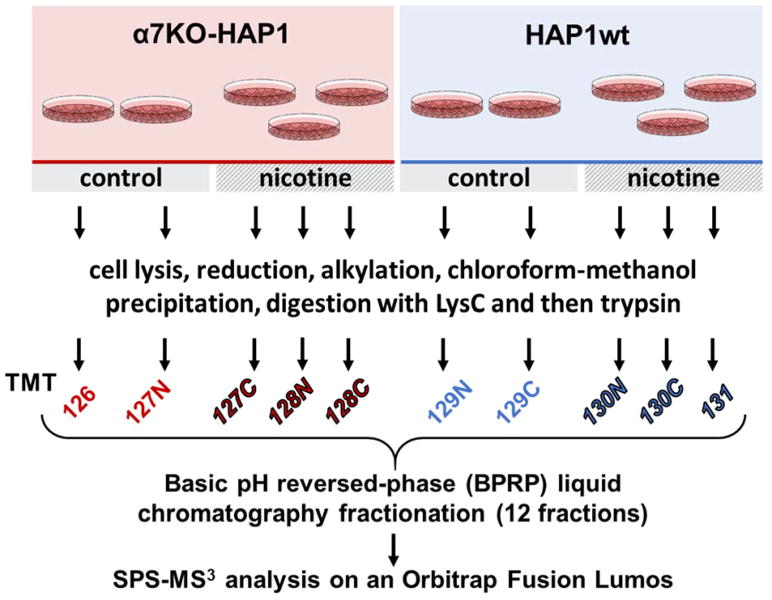
Figure 1. Experimental overview of the SPS-MS3 strategy.
Two cell types, α7KO-HAP1 and HAP1wt, were propagated and designated cultures were mock or nicotine-treated. Proteins were extracted and then digested with LysC and trypsin. The resulting peptides were labeled with TMT, pooled, and fractionated via basic pH reversed-phase (BPRP) high performance liquid chromatography (HPLC) prior to SPS-MS3 analysis on an Orbitrap Fusion Lumos mass spectrometer.