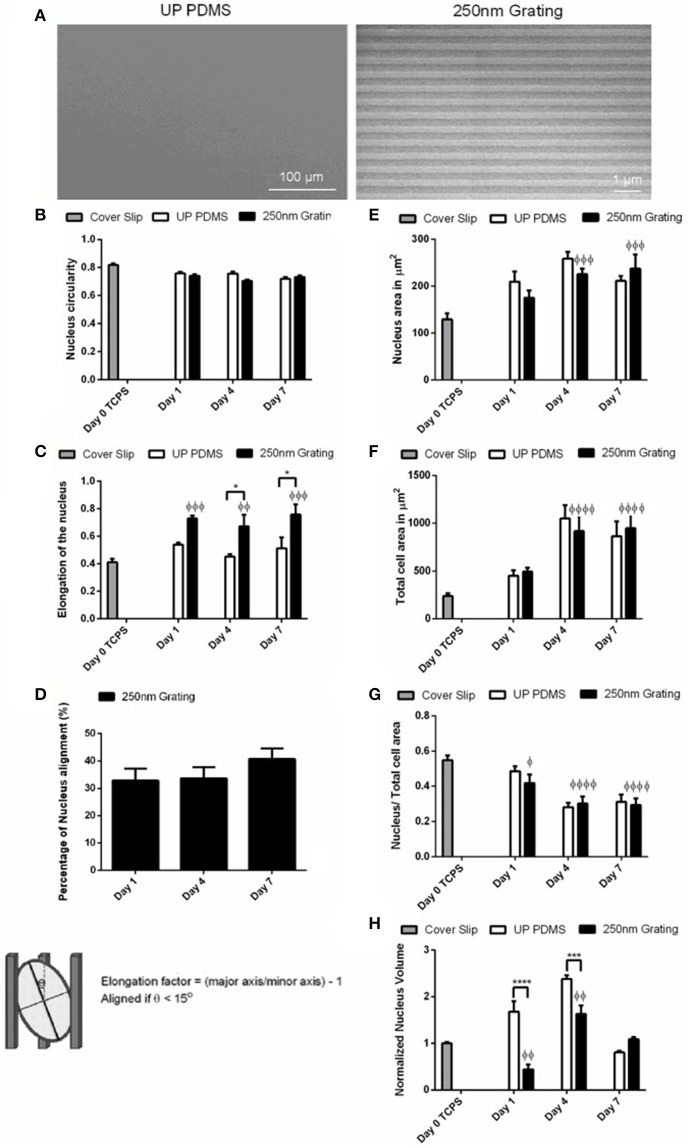
Figure 2.
(A) Scanning electron microscopy (SEM) image of unpatterned PDMS substrates (control) and 250 nm gratings. Temporal changes of the (B) nucleus circularity (comparison of replica mean values, N = 3), (C) nucleus elongation (comparison of replica mean values, N = 4), (D) nucleus alignment (comparison of experimental replicas, N = 3), (E) area of nucleus (comparison of experimental replicas, N = 3), (F) total cell area (comparison of experimental replicas, N = 3), (G) the ratio of nucleus area to total cell area (comparison of experimental replicas, N = 3), (H) normalized nucleus volume against Cover Slip control (comparison of experimental replicas, N = 4) were measured on hESCs grown on unpatterned and nano-patterned substrates during differentiation. At least 50 cells were analyzed in each of the experimental replica. *Represents statistical significance with p ≤ 0.05 against unpatterned PDMS, ***Represents statistical significance with p ≤ 0.001 against unpatterned PDMS, ****p ≤ 0.0001 against unpatterned PDMS, φp ≤ 0.05 against Cover Slip control, φφp ≤ 0.01 against Cover Slip control, φφφp ≤ 0.001 against Cover Slip control, and φφφφp ≤ 0.0001 against Cover Slip control. Data are shown as average ± SEM.