Abstract
Cholera, caused by the Gram-negative bacterium Vibrio cholerae, has ravaged humanity from time immemorial. Although the disease can be treated using antibiotics along with administration of oral rehydration salts and controlled by good sanitation, cholera is known to have produced mayhems in ancient times when little was known about the pathogen. By the 21st century, ample information about the pathogen, its epidemiology, genetics, treatment and control strategies was revealed. However, there is still fear of cholera outbreaks in developing countries, especially in the wake of natural calamities. Studies have proved that the bacterium is mutating and evolving, out-competing all our efforts to treat the disease with previously used antibiotics and control with existing vaccines. In this review, the major scientific insights of cholera research are discussed. Considering the important role of biofilm formation in the V. cholerae life cycle, the vast availability of next-generation sequencing data of the pathogen and multi-omic approach, the review thrusts on the identification of suitable biofilm-inhibiting targets and the discovery of anti-biofilm drugs from nature to control the disease.
Keywords: Cholera, integrons, multi-omic approach, rapid diagnostic tests, serotypes, Vibrio cholerae, whole genome
Introduction
Cholera (meaning being ‘a gutter’) is the right name for the disease, caused by the ingestion of contaminated water. The causative organism is a comma-shaped, flagellated bacterium, Vibrio cholerae. From 1817, there have been seven pandemics of cholera till date. High mortality rate and its spread in all major continents such as Asia, America, Europe and Africa make it important for research. The Indian subcontinent is considered to be the homeland of cholera, and from here, it had spread rapidly to other countries of the world. Symptoms that distinguish cholera from other diarrhoeal illnesses are the typical rice watery stool and severe vomiting that can lead to dehydration and death within 48 h if left untreated. According to the World Health Organization (WHO) report, 3.5 million people get infected and 100,000-120,000 people die due to cholera each year in the developing countries1. In 2014, the case fatality due to cholera was 1.17 per cent in 42 countries, which represented a 47 per cent increase as compared to 20131. Cholera pandemics are ranked sixth amongst the world's worst ten pandemics. The huge death rate is particularly reported by countries that suffer from poor sanitation facilities and lack of proper water distribution systems. Cross-border cases of cholera have been a serious issue in the Sub-Saharan African countries since the past many years raising the case-fatality rate to 5 per cent2. School-going children and children below five years are most affected in cholera-endemic countries. Cholera cases and deaths in African and South Asian countries have accounted for 99 per cent of the total cholera cases worldwide2.
Serotypes and biotypes
Over 200 serogroups of V. cholerae have been recognized so far, and among them, only a few produce the cholera toxin (CT), which is responsible for the characteristic clinical symptoms of the disease. The most common serogroups are O1 and O139 which cause epidemic cholera. The O1 serogroup is further classified into Ogawa, Inaba and Hikojima based on the mutation on wbeT gene and type of somatic O antigen formed. Generally, the non-O1/non-O139 strains do not cause the disease, but there have been rare cases of diarrheal infection caused by these3. In non-CT-producing V. cholerae, virulence factors such as heat-stable enterotoxin (Stn), haemolysin (Hly A), repeat in toxin (RTX) and type 3 secretion systems play major roles in causing infections4. Serogroup O1 is further subdivided into Classical and El Tor biotypes based on several phenotypic properties5. The classical strains produce larger amounts of CT than the prototype El Tor. The difference between classical and El Tor strains of V. cholerae is primarily based on phenotypic traits such as susceptibility to polymyxin B, chicken cell (erythrocytes) agglutination, haemolysis of sheep erythrocytes, Voges-Proskauer test, and phage susceptibilities where El Tor strains are susceptible to bacteriophage V but are resistant to bacteriophage IV; similarly, classical strains show the reverse characteristics in phage typing which is a gold standard test to differentiate classical and El Tor strains. Ogawa and Inaba serotypes are found to be common both in the classical and El Tor biotypes. V. cholerae belonging to the serogroup O139 was identified as a derivative of the serogroup O1 El Tor6. Till today, O1-specific antiserum is the best way to identify V. cholerae in the field. Major differences between El Tor and classical biotypes are also observed at the molecular level. Single nucleotide polymorphisms (SNPs) have been observed between the biotypes in the B subunit of CT (ctxB), toxin-coregulated pili (tcpA) and rtx. Previously, methods for identification of the pathogen were restricted to conventional biochemical and serological techniques. Later on, advancements in genomics and transcriptomics have helped us understand the pathogen better at the molecular level, however, the number of cases and high mortality rate caused by the pathogen still pertain in resource-poor countries. This makes cholera a disease that still deserves attention. In this review, the major scientific insights of cholera research are discussed with a view to develop an update of events to the scientific community in the concerned field.
Environmental reservoirs
The aquatic environments, including fresh, marine and brackish water bodies, have been identified as the natural reservoirs for cholera pathogen. A definite pattern of regular seasonal resurgence is characteristic of cholera7. Although studies have proven the existence of vibrios in association with copepods, shellfish, algae and in biofilm stage8, a clear and established understanding about the pathogen's inter-epidemic reservoir is not yet known. It has been observed that there are cholera outbreaks in September-October after a phytoplankton and zooplankton bloom in the water bodies of Bangladesh, a major epidemic-prone area8. Colwell et al9 demonstrated that a simple method of preventing the disease using inexpensive cloth (sari) folded four to eight times could greatly reduce the V. cholerae attached to plankton and also in biofilm state, thereby cutting down cholera cases drastically. It has also been shown that the bacterium enters into a dormant phase which is a viable but non-culturable (VBNC) state, accounting for why it could not be found in between epidemics using culture-dependent techniques10. Apart from planktons, V. cholerae also colonizes soft turtles and marine fishes which can be attributed as one of the major reasons for global dissemination of the pathogen11. Molecular subtyping of V. cholerae strains isolated from patient's stool samples and these soft turtles and fishes has shown high similarity, suggesting that these could serve as vehicles for the pathogen. Furthermore, this increases the incidence of cholera cases in those countries where sea foods are eaten raw or half cooked11. Cholera outbreaks have been reported in Vietnam after consumption of iced tea, showing the ability of V. cholerae to survive at low temperature and revive back in suitable conditions12.
History of cholera and Vibrio cholerae
Although there had been recorded cases of cholera in the 16th century, the knowledge about the causative organism improved only after two centuries. The first cholera case was well documented from India in 1563 by Garcia de Orta, a Portuguese physician13. Initially, it was believed that cholera was caused by bad air, arising from decayed organic matter or miasmata. Even after the invention of the microscope in 1660, the cause of cholera remained unknown to physicians of that time. It was in 1854, an Italian Anatomist Dr Filippo Pacini linked V. cholerae to the disease cholera by observing the pathogen in the intestine of corpses of the cholera epidemic at Florence13. With these observations, he described the disease as a massive loss of fluid and electrolytes from intestinal mucosa due to the bacteria and recommended the intravenous injection of saline in extreme cases of dehydration which went unnoticed until many years after his death14,15. Another major event in 1854 was that John Snow announced polluted water supply as the major cause for the spread of cholera in London16. Until then, the importance of clean water for human use and proper sanitation to prevent diarrhoeal diseases were not known. Snow's classical epidemiological research on the London cholera outbreak has helped in understanding the potential role of the contaminated water supply through public water pump on Broad Street as the source of cholera16. Through the epidemiological studies the first proofs to challenge the ‘miasma theory’ emanated. Later, only after two more lethal pandemics of cholera quivering the world from 1841 to 1875, Koch17 isolated the bacterium as a pure culture in 1883. The major events of cholera during 16th to 18th century and 19th century are summarized in Figure (A) and (B), respectively. At the end of the 19th century, there was a surge of vaccine trials in many cholera-stricken nations21,22. The vaccine trials existed over many decades, but the disease could not be contained, and the world saw the fifth and the sixth pandemics of cholera seizing many lives.
Figure.
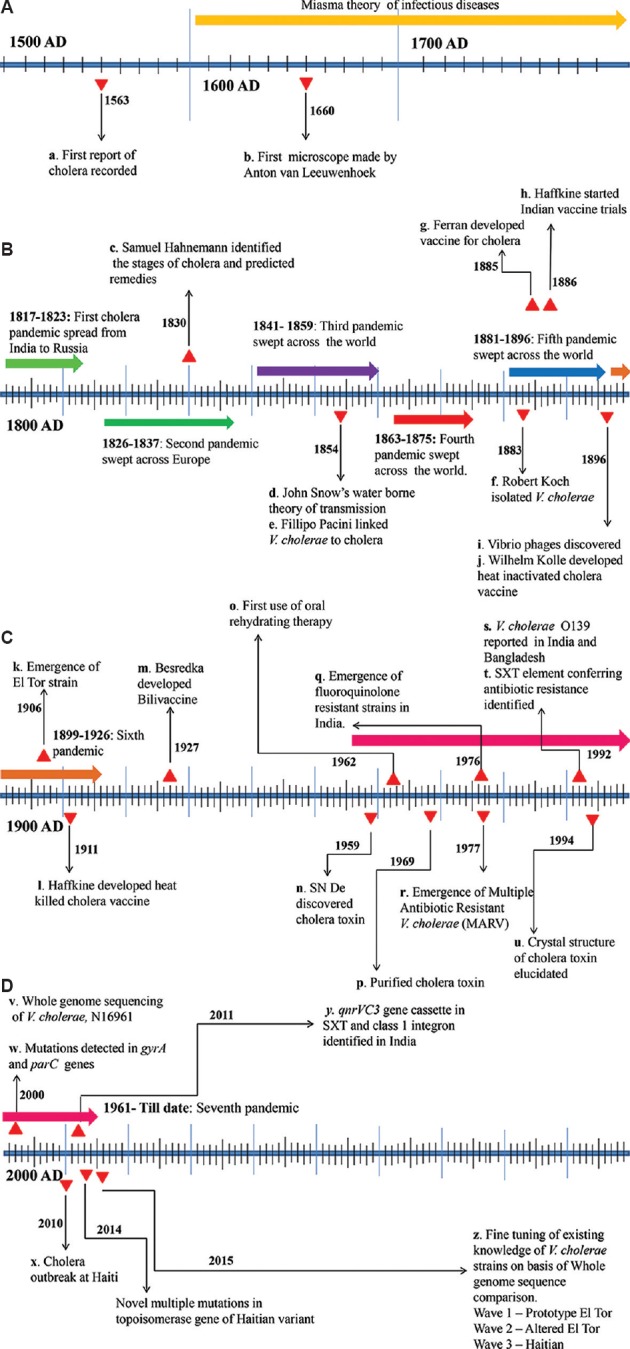
Timeline showing the major events and breakthroughs in cholera research. (A) 16th to 18th century, (B) 19th century, (C) 20th century, (D) 21st century till 2015 are represented.
Source: Refs. a13; b18; c19; d16; e14,15; f17; g21; h22; i20; j21; k27; l22; m21; n23; o29; p30; q25; r31; s28; t32; u33; v39; w44; x65; y66; z67.
The 20th century witnessed major discoveries in cholera research. In 1959, De, in Kolkata, India, identified CT to be responsible for the abysmal symptoms of cholera23. He performed experiments in rabbit ileum to replicate the disease and established CT as an enterotoxin23. This was a turning point in cholera research as it helped in understanding the pathogenicity of V. cholerae. Since then, there have been extensive studies on this pathogen, which led to better knowledge about the natural habitat, mode of infection, virulence and transmission of the bacteria. These discoveries modified the preventive measures used to control the disease. However, overcoming all discoveries, the pathogen evolved mechanisms to persist in the environment as well as withstand antimicrobial treatment by acquiring genes that conferred resistance. It is estimated that 40 per cent of the V. cholerae genes is part of mobile genetic elements which can be transferred from one strain to another distributing drug resistance24,25,26. Unlike the first six pandemics, the seventh pandemic was caused by the El Tor strains27. These new strains which were less virulent compared to the classical strains had the advantage of environmental persistence. In 1992, a novel strain of V. cholerae with epidemic potential was isolated from south India. It was designated as V. cholerae serogroup O139 Bengal since the first outbreak of the disease was reported from the coastal areas of the Bay of Bengal28. Since 1995, there have been variations in the clinical isolates of V. cholerae such that El Tor strains have been found to produce CT of the classical biotype. These strains are capable of being dominant in transmission as well as in virulence. One major development of 20th century was the identification of oral rehydration therapy to replenish lost fluid in cholera patients29. The milestones of cholera research in the 20th century are given in Figure (C).
Modern façade of Vibrio cholerae through era of multi-Omics
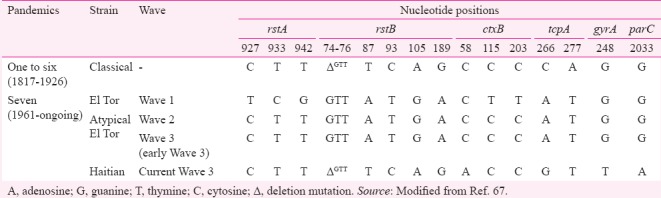
The development of molecular techniques such as 16S-23S intergenic spacer region amplification made it easier to differentiate V. cholerae from other vibrios34. The genomic analysis of V. cholerae revolved around understanding the differences between the two biotypes, classical and El Tor. Septaplex and multiplex polymerase chain reactions have been developed for detecting major virulence and resistance genes, which could be used to differentiate these biotypes35,36. Important variations in the genetic organization of the evolving strains of V. cholerae are reported in TCP and CT. Such genetic changes would have played significant roles in the difference in their pathogenicity37,38. The genes that encode CT (ctxAB) exist within the genome of a lysogenic filamentous bacteriophage CTXφ, first discovered by Waldor and Mekalanos20. The CTXφ is integrated at one chromosome or both the chromosomes within El Tor and classical, respectively. The major differences between the biotypes of V. cholerae at the CTX gene cluster and fine tuning of V. cholerae strains based on the type of CtxB they harbour are described in the Table.
Table.
Major differences between virulence, regulatory and quinolone resistance-determining region of Vibrio cholerae epidemic strains at nucleotide level (A, G, T and C)
Applications of next-generation sequencing
Whole genome sequencing of V. cholerae by Heidelberg et al39 was a milestone in understanding the pathogen in the genomic level. It was revealed that V. cholerae possesses two circular chromosomes of 2,961,146 bp and 1,072,314 bp collectively coding for 3885 open reading frames. The majority of genes for essential cell functions and pathogenicity are located on the large chromosome and small chromosome contains majorly hypothetical genes and genes usually present in the plasmids39. V. cholerae genome sequence initiated the understanding of how an environmental non-toxigenic bacterium emerged and evolved to become a significant human bacterial pathogen. It is through changes in global patterns of transcription that V. cholerae adapts to different environmental conditions. These transcriptional changes alter the organism's protein repertoire which in turn changes its metabolic status40. Thus, information on global changes in the V. cholerae transcriptome in both in vitro as well as in vivo conditions had been helpful in the analysis of the adaptive processes. It also provided potential insight for therapeutics. A notable difference is seen in the genes being transcribed in in vivo and in vitro conditions. Finally, V. cholerae comparative genomics was also studied using microarray41. Global proteome profiles of pandemic strains of V. parahaemolyticus and V. cholerae in planktonic and biofilm stage have provided an understanding of the importance of genes such as mannose-sensitive haemagglutinin and chitin-regulated pilus in biofilm formation42. The outer membrane vesicle of V. cholerae has been studied through high-throughput lipidomics43. All these studies have informed that the genomic plasticity of the pathogen is very high and acquisition of new genes in V. cholerae enhances its chances of survival by allowing growth in a previously hostile environment. Notable genetic changes such as transposable elements and site-specific recombination systems such as integrons and super-integrons have been identified in V. cholerae. These genetic elements can mobilize genes from one replicon to another, thus establishing conditions for interspecies and intraspecies gene transfer. The current variants have revealed a gyrA mutation for quinolone resistance and also possessed integrating conjugative elements that confer multiple drug resistance, to trimethoprim, streptomycin and chloramphenicol44,45. The currently circulating strains also possessed multiple novel mutations in the topoisomerase gene, which is further attributed to multidrug resistance46. Although genomics has revealed a lot about the genetic variations in V. cholerae, each new outbreak instigates a fresh demand for more studies as the pathogen is still evolving.
Fighting the disease
At present, cholera is managed with oral rehydration solution (ORS) to correct dehydration and supplemented with antibiotics in extreme cases. ORS is one of the greatest discoveries in cholera prophylaxis which is cheap and easy to prepare even in the most remote areas. The currently used cholera vaccines do not give complete protection against the disease for a lifetime. The WHO recommends the use of cholera vaccines only in combination with other preventive measures among those at high risks. Shanchol® (manufactured by Shantha Biotech in India) and Dukoral® (manufactured by SBL Vaccines) are the two WHO prequalified oral cholera vaccines used against cholera47. In 2016, the WHO approved a single-dose live oral cholera vaccine, Vaxchora® in the United States for adults aged 18-64 yr travelling to a cholera-endemic area48. Several live attenuated cholera vaccines that have the ability of providing long-term protection with a single dose are under development, of which none are expected to be marketed within the following few years. Mass vaccination against cholera is not recommended in cholera-endemic countries. However, cholera vaccination in high-risk population such as children below five, school-going children, pregnant women, old age groups and immunocompromised individuals is a requisite in such countries.
Rapid diagnostic tests
Simple and reliable methods for detecting V. cholerae from patients’ stool samples and the environment are of great value for epidemiologists, clinicians and health officials in instigating control measures before an outbreak. Rapid diagnostic tests (RDTs) require minimum laboratory infrastructure, less time and basic technical skills as compared to the routine culture techniques. Currently, more than 20 RDTs are marketed which have been field tested by the WHO and Global Task Force on Cholera Control (GTFCC) Surveillance Laboratory Working Group49. Efficacy, mode of detection, structure of RDTs and their use differ from region to region. Cholera RDTs in the market are either lateral flow units or dipsticks and work by detecting either CT50,51 or antigenic lipopolysaccharide (LPS) of V. cholerae52. Some of the RDTs such as Sensitive Membrane Antigen Rapid Test (SMART) II (developed by New Horizons Diagnostics Corporation, Columbia, MD) utilizes a calorimetric gold-based immunoassay with monoclonal antibody specific for A antigen of O1 LPS of either V. cholerae O1 or O13953. There are a few RDTs that utilize the polyclonal antibody-based systems that can detect both V. cholerae O1 and O139 with the same kit. A few commonly encountered problems in using RDTs are that (i) these tests may detect both viable and non-viable bacteria. In addition, a shortly vaccinated person's stool sample gives positive results as many of the vaccines have LPS as its major component, and (ii) some RDTs show cross-reactivity to closely related species of microorganisms. However, Crystal VC (Span Diagnostic, Surat, India) has a high sensitivity of 92-100 per cent and has been used for several field studies in India54.
Identification of Anti-biofilm targets and compounds
As V. cholerae has dual life cycle both within the host body and in aquatic environments, the bacterium has been used as a model organism to understand the molecular mechanisms and signals underlying biofilm formation, assembly and dispersal for last 20 years55. Biofilm assembly is initiated by change in global transcriptome profile downregulating genes for motility and chemotaxis and upregulating genes important for the synthesis of its extracellular exopolysaccharide matrix. The major exopolysaccharide Vibrio polysaccharide is encoded by a cassette of vps genes, which is in turn controlled by rpoS. Further it has been demonstrated that c-di-GMP (cyclic diguanylate monophosphate) signalling regulates transition of cells between the planktonic and biofilm state56. The increase and decrease of the levels of c-di-GMP within the V. cholerae cell are regulated by diguanylate cyclase (DGC) and phosphodiesterases, respectively. These regulations for biofilm development and dispersal at the genetic level are in turn associated with the environmental cues the bacteria receive. Among the conditions that affect the biofilm development are temperature, pH, O2 levels, hydrodynamics, osmolarity, presence of specific ions, nutrients and factors derived from the biotic environment57. The role of biofilms in the environmental persistence, dissemination, tolerance to antibiotics and transmission of V. cholerae has been well established. Increasing cholera outbreaks spanning different continents and emergence of multidrug resistance in V. cholerae necessitate an urgent need to look for alternative strategies to combat the infections by reducing virulence rather than by killing the bacteria. Thus, discovering potential anti-biofilm compounds against V. cholerae and finding their anti-biofilm targets are important.
A single biofilm inhibitor can theoretically control both virulence, biofilm formation and dispersal of the pathogen. Anti-biofilm drugs from nature, such as phytochemicals, frog skin peptides, human breast milk-derived small molecules and probiotic microbes that have anti-biofilm activity against vibrios, are fast-gaining popularity as these have fewer side effects. In addition, the pathogen's ability to develop resistance against these natural products is considered to be limited58,59,60. Probiotic strains, such as Ruminococcus obeum that are non-infectious and indigenous to human gut, are known to limit colonization of V. cholerae and other enteric pathogens61. Transcriptomic and proteomic approaches to identify conserved biofilm inhibiting targets are also promising strategies to strengthen the fight against cholera.
The metagenomic approach in cholera research has produced unexpected results, showing the presence of V. cholerae in the gut of healthy children, which requires detailed investigation62. The presence of V. cholerae in the VBNC state has been understood by metagenomics63. In spite of having substantial knowledge about the pathogen in the 21st century and novel strategies developed to fight the disease, there is a dearth of access to an effective therapy for cholera in developing countries64. Cholera research is expanding fast in the 21st century, as represented in Figure (D).
Conclusion
Today, the perception of V. cholerae as an uncontrollable ferocious pathogen lurking in polluted waters, causing large-scale morbidity and mortality, is changed. We have now several effective measures in controlling the infection and spread of the disease. However, V. cholerae prevails in the environment during inter-epidemic periods unnoticed out-competing our efforts to control the disease during natural disasters such as floods and earthquakes that lead to unhygienic environment and overcrowding. The best we could do is to understand the evolving strains and avoid another major outbreak that has epidemic and pandemic potential. Although there have been a few initiatives to identify a potential biofilm inhibiting drug target in the pathogen60,68, there is still a vacuum in this area. The variations and difference in pathogenicity of the evolved and emerging strains are to be studied. To date, 26 complete genomes and more than 220 draft genomes of V. cholerae are available in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/genome/genomes/505). A detailed spatiotemporal investigation of the genomic diversity of outbreak strains is warranted to study the evolutionary adaptation and transmission disparity of the pathogen. An interpretation compiling the transcriptomic, proteomic and metabolomic data available in the respective databanks to identify a suitable drug target in the Vibrionaceae family at large is not an unattainable task. Further, updated knowledge on the history of cholera and pathogenicity of V. cholerae in relation to its epidemiology and evolution is essential. Hence, the coordination of all cholera research groups and strong establishment of international networking is warranted for the battle against cholera.
Acknowledgment
Authors thank Prof. M. Radhakrishna Pillai, Director, Rajiv Gandhi Centre for Biotechnology (RGCB), Thiruvananthapuram, India, for the facilities provided.
Footnotes
Financial support & sponsorship: The first author (LN) received the INSPIRE fellowship from Department of Science & Technology (DST), Government of India, New Delhi, India
Conflicts of Interest: None.
References
- 1.Cholera. 2014. Wkly Epidemiol Rec. 2015;90:517–28. [PubMed] [Google Scholar]
- 2.Bwire G, Mwesawina M, Baluku Y, Kanyanda SSE, Orach CG. Cross-border cholera outbreaks in Sub-Saharan Africa, the mystery behind the silent illness: What needs to be done? PLoS One. 2016;11:e0156674. doi: 10.1371/journal.pone.0156674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen YT, Tang HJ, Chao CM, Lai CC. Clinical manifestations of non-O1 Vibrio cholerae infections. PLoS One. 2015;10:e0116904. doi: 10.1371/journal.pone.0116904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty S, Mukhopadhyay AK, Bhadra RK, Ghosh AN, Mitra R, Shimada T, et al. Virulence genes in environmental strains of Vibrio cholerae. Appl Environ Microbiol. 2000;66:4022–8. doi: 10.1128/aem.66.9.4022-4028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svennerholm AM, Holmgren J. Identification of Escherichia coli heat-labile enterotoxin by means of a ganglioside immunosorbent assay (GM1-ELISA) Curr Microbiol. 1978;1:19–23. [Google Scholar]
- 6.Raychoudhuri A, Mukhopadhyay AK, Ramamurthy T, Nandy RK, Takeda Y, Nair GB, et al. Biotyping of Vibrio cholerae O1: Time to redefine the scheme. Indian J Med Res. 2008;128:695–8. [PubMed] [Google Scholar]
- 7.Wu X, Lu Y, Zhou S, Chen L, Xu B. Impact of climate change on human infectious diseases: Empirical evidence and human adaptation. Environ Int. 2016;86:14–23. doi: 10.1016/j.envint.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Huq A, Sack RB, Nizam A, Longini IM, Nair GB, Ali A, et al. Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl Environ Microbiol. 2005;71:4645–54. doi: 10.1128/AEM.71.8.4645-4654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colwell RR, Huq A, Islam MS, Aziz KM, Yunus M, Khan NH, et al. Reduction of cholera in Bangladeshi villages by simple filtration. Proc Natl Acad Sci U S A. 2003;100:1051–5. doi: 10.1073/pnas.0237386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alam M, Sultana M, Nair GB, Siddique AK, Hasan NA, Sack RB, et al. Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc Natl Acad Sci U S A. 2007;104:17801–6. doi: 10.1073/pnas.0705599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Yan M, Gao H, Lu X, Kan B. Vibrio cholerae colonization of soft-shelled turtles. Appl Environ Microbiol. 2017;83:pii : e00713–7. doi: 10.1128/AEM.00713-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen TV, Pham QD, Do QK, Diep TT, Phan HC, Ho TV, et al. Cholera returns to Southern Vietnam in an outbreak associated with consuming unsafe water through iced tea: A matched case-control study. PLoS Negl Trop Dis. 2017;11:e0005490. doi: 10.1371/journal.pntd.0005490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen S. Indian cholera: A Myth. IJHS. 2012;47:345–74. [Google Scholar]
- 14.Nardi MG. Discovery of Vibrio cholerae by Filippo Pacini, of Pistoia, established in the initial phases of microbiological thought and judged after a century. Minerva Med. 1954;45:1024–9. [PubMed] [Google Scholar]
- 15.Subba Rao M, Howard-Jones N. Original observations of Filippo Pacini on Vibrio cholera. Bull Indian Inst Hist Med Hyderabad. 1978;8:32–8. [PubMed] [Google Scholar]
- 16.Snow J. On the Mode of Communication of Cholera. London: John Churchill; 1855. [Google Scholar]
- 17.Koch R. An address on cholera and its Bacillus. Br Med J. 1884;2:403–7. doi: 10.1136/bmj.2.1235.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egerton FN. Leeuwenhoek as a founder of animal demography. J Hist Biol. 1967;1:1–22. [Google Scholar]
- 19.Hahnemann S. Appeal to Thinking Philanthropist Respecting the Mode of Propagation of Asiatic Cholera. The Lesser Writings of Samuel Hahnemann. 1831:753–63. [Google Scholar]
- 20.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–4. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 21.Kabir S. Cholera vaccines: The current status and problem. Rev Med Microbiol. 2005;16:101–16. [Google Scholar]
- 22.Hawgood BJ. Waldemar Mordecai Haffkine, CIE (1860-1930): Prophylactic vaccination against cholera and bubonic plague in British India. J Med Biogr. 2007;15:9–19. doi: 10.1258/j.jmb.2007.05-59. [DOI] [PubMed] [Google Scholar]
- 23.De SN. Enterotoxicity of bacteria-free culture-filtrate of Vibrio cholerae. Nature. 1959;183:1533–4. doi: 10.1038/1831533a0. [DOI] [PubMed] [Google Scholar]
- 24.Jesudason MV, Saaya R. Resistance of Vibrio cholerae 01 to nalidixic acid. Indian J Med Res. 1997;105:153–4. [PubMed] [Google Scholar]
- 25.Mukhopadhyay AK, Basu I, Bhattacharya SK, Bhattacharya MK, Nair GB. Emergence of fluoroquinolone resistance in strains of Vibrio cholerae isolated from hospitalized patients with acute diarrhea in Calcutta, India. Antimicrob Agents Chemother. 1998;42:206–7. doi: 10.1128/aac.42.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sack DA, Lyke C, McLaughlin C, Suwanvanichkij V. Geneva: World Health Organization; 2001. Antimicrobial resistance in shigellosis, cholera and campylobacteriosis. [Google Scholar]
- 27.Hu D, Liu B, Feng L, Ding P, Guo X, Wang M, et al. Origins of the current seventh cholera pandemic. Proc Natl Acad Sci U S A. 2016;113:E7730–9. doi: 10.1073/pnas.1608732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramamurthy T, Garg S, Sharma R, Bhattacharya SK, Nair GB, Shimada T, et al. Emergence of novel strain of Vibrio cholerae with epidemic potential in Southern and Eastern India. Lancet. 1993;341:703–4. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 29.Ruxin JN. Magic bullet: The history of oral rehydration therapy. Med Hist. 1994;38:363–97. doi: 10.1017/s0025727300036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkelstein RA, LoSpalluto JJ. Pathogenesis of experimental cholera. Preparation and isolation of choleragen and choleragenoid. J Exp Med. 1969;130:185–202. doi: 10.1084/jem.130.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faruque AS, Alam K, Malek MA, Khan MG, Ahmed S, Saha D, et al. Emergence of multidrug-resistant strain of Vibrio cholerae O1 in Bangladesh and reversal of their susceptibility to tetracycline after two years. J Health Popul Nutr. 2007;25:241–3. [PMC free article] [PubMed] [Google Scholar]
- 32.Thungapathra M, Amita, Sinha KK, Chaudhuri SR, Garg P, Ramamurthy T, et al. Occurrence of antibiotic resistance gene cassettes aac(6’)-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob Agents Chemother. 2002;46:2948–55. doi: 10.1128/AAC.46.9.2948-2955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merritt EA, Sarfaty S, van den Akker F, L’Hoir C, Martial JA, Hol WG. Crystal structure of cholera toxin B-pentamer bound to receptor GM1 pentasaccharide. Protein Sci. 1994;3:166–75. doi: 10.1002/pro.5560030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chun J, Huq A, Colwell RR. Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl Environ Microbiol. 1999;65:2202–8. doi: 10.1128/aem.65.5.2202-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, et al. Development and evaluation of a Multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol. 1998;20:201–7. doi: 10.1111/j.1574-695X.1998.tb01128.x. [DOI] [PubMed] [Google Scholar]
- 36.Mantri CK, Mohapatra SS, Ramamurthy T, Ghosh R, Colwell RR, Singh DV, et al. Septaplex PCR assay for rapid identification of Vibrio cholerae including detection of virulence and int SXT genes. FEMS Microbiol Lett. 2006;265:208–14. doi: 10.1111/j.1574-6968.2006.00491.x. [DOI] [PubMed] [Google Scholar]
- 37.Häse CC, Mekalanos JJ. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A. 1998;95:730–4. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukhopadhyay AK, Chakraborty S, Takeda Y, Nair GB, Berg DE. Characterization of VPI pathogenicity island and CTXphi prophage in environmental strains of Vibrio cholerae. J Bacteriol. 2001;183:4737–46. doi: 10.1128/JB.183.16.4737-4746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–83. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beyhan S, Tischler AD, Camilli A, Yildiz FH. Differences in gene expression between the classical and el tor biotypes of Vibrio cholerae O1. Infect Immun. 2006;74:3633–42. doi: 10.1128/IAI.01750-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, Mekalanos JJ, et al. Comparative genomic analysis of Vibrio cholerae: Genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci U S A. 2002;99:1556–61. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dharmaprakash A, Mutt E, Jaleel A, Ramanathan S, Thomas S. Proteome profile of a pandemic Vibrio parahaemolyticus SC192 strain in the planktonic and biofilm condition. Biofouling. 2014;30:729–39. doi: 10.1080/08927014.2014.916696. [DOI] [PubMed] [Google Scholar]
- 43.Kulkarni HM, Jagannadham MV. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology. 2014;160:2109–21. doi: 10.1099/mic.0.079400-0. [DOI] [PubMed] [Google Scholar]
- 44.Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–4. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 45.Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK, et al. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother. 2001;45:2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Divya MP, Sivakumar KC, Sarada Devi KL, Remadevi S, Thomas S. Novel multiple mutations in the topoisomerase gene of Haitian variant Vibrio cholerae O1. Antimicrob Agents Chemother. 2014;58:4982–3. doi: 10.1128/AAC.03189-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez AL, Gonzales ML, Aldaba JG, Nair GB. Killed oral cholera vaccines: History, development and implementation challenges. Ther Adv Vaccines. 2014;2:123–36. doi: 10.1177/2051013614537819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cabrera A, Lepage JE, Sullivan KM, Seed SM. Vaxchora: A single-dose oral cholera vaccine. Ann Pharmacother. 2017;51:584–9. doi: 10.1177/1060028017698162. [DOI] [PubMed] [Google Scholar]
- 49.Ontweka LN, Deng LO, Rauzier J, Debes AK, Tadesse F, Parker LA, et al. Cholera rapid test with enrichment step has diagnostic performance equivalent to culture. PLoS One. 2016;11:e0168257. doi: 10.1371/journal.pone.0168257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Almeida RJ, Hickman-Brenner FW, Sowers EG, Puhr ND, Farmer JJ, 3rd, Wachsmuth IK, et al. Comparison of a latex agglutination assay and an enzyme-linked immunosorbent assay for detecting cholera toxin. J Clin Microbiol. 1990;28:128–30. doi: 10.1128/jcm.28.1.128-130.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yam WC, Lung ML, Ng MH. Evaluation and optimization of a latex agglutination assay for detection of cholera toxin and Escherichia coli heat-labile toxin. J Clin Microbiol. 1992;30:2518–20. doi: 10.1128/jcm.30.9.2518-2520.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carillo L, Gilman RH, Mantle RE, Nunez N, Watanabe J, Moron J, et al. Rapid detection of Vibrio cholerae O1 in stools of Peruvian cholera patients by using monoclonal immunodiagnostic kits. Loyaza Cholera Working Group in Peru. J Clin Microbiol. 1994;32:856–7. doi: 10.1128/jcm.32.3.856-857.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hasan JA, Huq A, Tamplin ML, Siebeling RJ, Colwell RR. A novel kit for rapid detection of Vibrio cholerae O1. J Clin Microbiol. 1994;32:249–52. doi: 10.1128/jcm.32.1.249-252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ley B, Khatib AM, Thriemer K, von Seidlein L, Deen J, Mukhopadyay A, et al. Evaluation of a rapid dipstick (Crystal VC) for the diagnosis of cholera in Zanzibar and a comparison with previous studies. PLoS One. 2012;7:e36930. doi: 10.1371/journal.pone.0036930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, et al. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–9. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valentini M, Filloux A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem. 2016;291:12547–55. doi: 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teschler JK, Zamorano-Sánchez D, Utada AS, Warner CJA, Wong GCL, Linington RG, et al. Living in the matrix: Assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol. 2015;13:255–68. doi: 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ammons MC, Copié V. Mini-review: Lactoferrin: A bioinspired, anti-biofilm therapeutic. Biofouling. 2013;29:443–55. doi: 10.1080/08927014.2013.773317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brackman G, Defoirdt T, Miyamoto C, Bossier P, Van Calenbergh S, Nelis H, et al. Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in Vibrio spp.by decreasing the DNA-binding activity of the quorum sensing response regulator LuxR. BMC Microbiol. 2008;8:149. doi: 10.1186/1471-2180-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Augustine N, Goel AK, Sivakumar KC, Kumar RA, Thomas S. Resveratrol - a potential inhibitor of biofilm formation in Vibrio cholerae. Phytomedicine. 2014;21:286–9. doi: 10.1016/j.phymed.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 61.Hsiao A, Ahmed AM, Subramanian S, Griffin NW, Drewry LL, Petri WA, Jr, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–6. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monira S, Nakamura S, Gotoh K, Izutsu K, Watanabe H, Alam NH, et al. Metagenomic profile of gut microbiota in children during cholera and recovery. Gut Pathog. 2013;5:1. doi: 10.1186/1757-4749-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Materna AC, Friedman J, Bauer C, David C, Chen S, Huang IB, et al. Shape and evolution of the fundamental niche in marine Vibrio. ISME J. 2012;6:2168–77. doi: 10.1038/ismej.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghosh A, Ramamurthy T. Antimicrobials & cholera: Are we stranded? Indian J Med Res. 2011;133:225–31. [PMC free article] [PubMed] [Google Scholar]
- 65.Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, et al. The origin of the Haitian cholera outbreak strain. N Engl J Med. 2011;364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar P, Thomas S. Presence of qnrVC3 gene cassette in SXT and class 1 integrons of Vibrio cholerae. Int J Antimicrob Agents. 2011;37:280–1. doi: 10.1016/j.ijantimicag.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 67.Kim EJ, Lee CH, Nair GB, Kim DW. Whole-genome sequence comparisons reveal the evolution of Vibrio cholerae O1. Trends Microbiol. 2015;23:479–89. doi: 10.1016/j.tim.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 68.Dharmaprakash A, Thandavarayan R, Joseph I, Thomas S. Development of broad-spectrum antibiofilm drugs: Strategies and challenges. Future Microbiol. 2015;10:1035–48. doi: 10.2217/fmb.15.14. [DOI] [PubMed] [Google Scholar]


