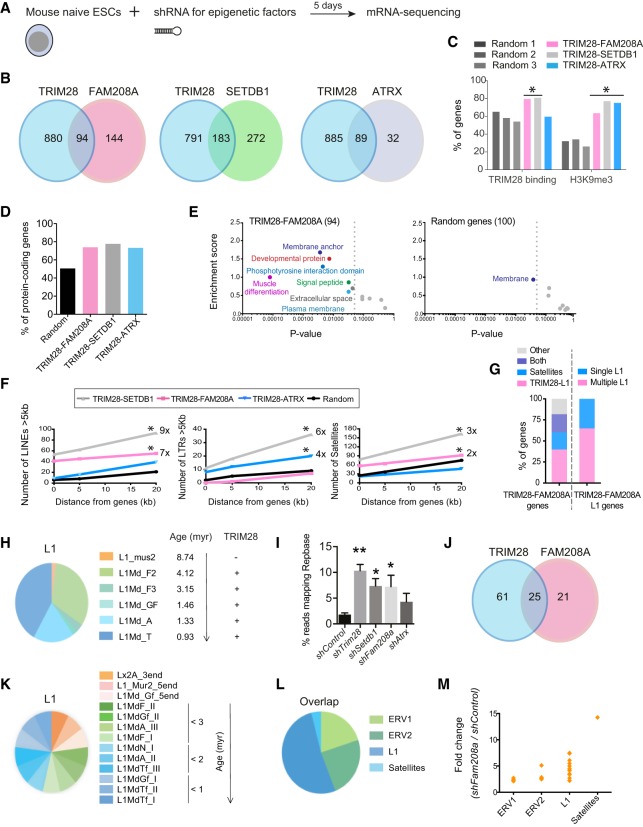
Figure 3.
TRIM28 and FAM208A exert nonredundant roles at evolutionarily young L1s and associated genes. (A) Naïve knockdown J1 mESCs were subject to mRNA-sequencing. Biological replicates were sequenced from three independent experiments. (B) Genes up-regulated >2× (where Padj ≤ 0.05) in each treatment group showing the overlap between groups. (C) The three gene sets or three random gene sets (the latter containing 100 per group) were examined for the presence of a TRIM28 or H3K9me3 peak within a radius of 20 kb. (D) Percentage of protein-coding genes in each group. (E) Gene ontology (DAVID analysis) of the 94 TRIM28-FAM208A repressed genes (left, seven gene clusters were enriched with P-values <0.05) and the 100 random genes (right). (F) UCSC Table Browser analysis showing the number of the stated repeats located within increasing distances (0, 5, and 20 kb) of the sets of genes. Significant gene sets are marked and the fold change relative to random genes at intersection (0 kb) is stated where different. (Left) TRIM28-FAM208A, P = 0.000025; TRIM28-SETDB1, P = 0.002540; (middle) TRIM28-ATRX, P = 0.010200; TRIM28-SETDB1, P = 0.035700; (right) TRIM28-FAM208A, P = 0.00643; TRIM28-SETDB1, P = 0.00428. (G) The percentage of TRIM28-FAM208A genes that contain the stated repeats (left bar) or the percentage of L1-containing TRIM28-FAM208A genes that contain multiple L1s (right bar). Only TRIM28-dependent L1s are considered (from the families L1Md_F, L1Md_F2, L1Md_F3, L1Md_A, and L1Md_T). (H) Full-length (>5 kb) L1 elements located within 20 kb of the TRIM28-FAM208A genes were classified according to family and mean age of that family and whether (+/−) they bind TRIM28. (I) The percentage of reads mapping Repbase within each treatment group is shown (n = 3, except for ATRX where n = 2). Error bars represent standard deviation or standard error (ATRX). (J) Venn diagram showing 25 repeat families are co-repressed by TRIM28 and FAM208A. They are defined as >2× up-regulated (P = <0.05) in both Trim28 and Fam208a-depleted cells. (K) All L1 families co-repressed by TRIM28 and FAM208A are classified here by name and age. (L) Proportion of repeats from each class that are co-repressed by TRIM28 and FAM208A. (M) The same repeat families as L, but here, their up-regulation in Fam208a-depleted cells is shown.