Abstract
Several common alleles in the oxytocin receptor gene (OXTR) are associated with altered brain function in reward circuitry in neurotypical adults and may increase risk for autism spectrum disorders (ASD). Yet, it is currently unknown how variation in the OXTR relates to brain functioning in individuals with ASD, and, critically, whether neural endophenotypes vary as a function of aggregate genetic risk. Here, for we believe the first time, we use a multi-locus approach to examine how genetic variation across several OXTR single-nucleotide polymorphisms (SNPs) affect functional connectivity of the brain’s reward network. Using data from 41 children with ASD and 41 neurotypical children, we examined functional connectivity of the nucleus accumbens (NAcc) – a hub of the reward network – focusing on how connectivity varies with OXTR risk-allele dosage. Youth with ASD showed reduced NAcc connectivity with other areas in the reward circuit as a function of increased OXTR risk-allele dosage, as well as a positive association between risk-allele dosage and symptom severity, whereas neurotypical youth showed increased NAcc connectivity with frontal brain regions involved in mentalizing. In addition, we found that increased NAcc-frontal cortex connectivity in typically developing youth was related to better scores on a standardized measure of social functioning. Our results indicate that cumulative genetic variation on the OXTR impacts reward system connectivity in both youth with ASD and neurotypical controls. By showing differential genetic effects on neuroendophenotypes, these pathways elucidate mechanisms of vulnerability versus resilience in carriers of disease-associated risk alleles.
INTRODUCTION
Autism spectrum disorders (ASD) are genetically complex neurodevelopmental disorders. Although hundreds of genes have been implicated in the etiology of ASD, recent evidence suggests that most genetic liability is conferred by inherited variations in single-nucleotide polymorphisms (SNPs) that are commonly distributed throughout the population.1,2 Findings of risk variants clustered within particular genes may suggest biological pathways likely to be altered in ASD, supporting an additive, polygenic model of autism risk whereby individuals who carry greater numbers of risk alleles are at increased risk for ASD and may present more severe symptomatology.3 However, thus far, studies relating common genetic variants to neurobiological functioning in ASD have focused on single SNPs. This approach fails to take advantage of genetic variability across multiple loci, which can be leveraged to parse the considerable neurobiological heterogeneity observed in ASD.
As social deficits are core features of the ASD phenotype, genes linked to variations in social behavior are logical targets for investigation. Allelic variations on the oxytocin receptor gene (OXTR) have been associated with increased rates of ASD,4 lower social responsiveness5 and increased severity of social deficits in individuals with ASD.5–7 Further evidence for the critical role of OXTR in social functioning comes from animal models demonstrating that dense expression of the oxytocin receptor in the nucleus accumbens (NAcc) is associated with social affiliative behaviors,8 reduces susceptibility to early adverse experiences (for example, neglect)9 and critically, that allelic variation in the OXTR is associated with variability in receptor expression specifically in brain areas important for social attachment, including the NAcc.10 Further, disruption of oxytocin receptor signaling in the NAcc inhibits social attachment11,12 and the formation of positive associations with social rewards.13 Together, these studies suggest that oxytocin receptor function in the NAcc is required to form associations with social rewards through its effects on neural excitability in reward-related brain regions.
Notably, a large body of work indicates that reward processing is altered at the neural level in individuals with ASD. Compared with neurotypical controls, individuals with ASD show aberrant brain activity to monetary rewards in several nodes of reward circuitry including the ventral striatum, anterior cingulate and prefrontal cortex,14–17 as well as reduced striatal activity to positive social rewards (that is, smiling faces).15,18 According to a leading theory of ASD, reduced ability to represent the reward-value of social stimuli results in poor motivation to engage in social interactions, decreasing opportunities for social learning and thus leading to social impairments.19,20 Importantly, imaging-genetics studies in neurotypical adults suggest that inheritance of certain OXTR alleles linked to poor social skills are related to altered functional activity, connectivity and volume of brain regions implicated in reward and social-emotional processing.21–23 However, no studies have examined how any OXTR SNPs relate to brain functioning in individuals with ASD and, importantly, whether neural endophenotypes vary as a function of carrying multiple OXTR risk alleles.
We addressed these questions in 41 youth with ASD and 41 typically developing (TD) matched controls (age range 9–17 years; Table 1) using resting-state functional magnetic resonance imaging to examine the relationship between four OXTR SNPs previously associated with ASD4,24–26 (rs53576, rs237887, rs1042778 and rs2254298) and functional connectivity in reward circuitry. Based on the animal and human work presented above, we hypothesized that greater OXTR genetic risk would be associated with reduced NAcc functional connectivity with other nodes of the reward system, particularly so in youth with ASD.
Table 1.
Subject characteristics
| Characteristic | TD subjects | ASD subjects | P-value |
|---|---|---|---|
| N | 41 | 41 | |
| Gender | 34 male | 37 male | |
| Handedness | 38 right-handed | 36 right-handed | |
| Age (years) | 13.11 (1.8) | 13.52 (2.2) | 0.36 |
| Full-scale IQ | 106.12 (10.2) | 104.54 (14.4) | 0.57 |
| Verbal IQ | 106.59 (11.0) | 103.24 (13.3) | 0.22 |
| Performance IQ | 105.51 (11.1) | 105.12 (14.4) | 0.89 |
| ADOS (comm+ soc) | 11.85 (4.63) | ||
| ADI (comm) total | 16.29 (4.41) | ||
| Mean absolute motion | 0.25 (0.16) | 0.26 (0.14) | 0.72 |
| Max absolute motion | 0.75 (0.60) | 0.78 (0.45) | 0.84 |
| Mean framewise displacement | 0.06 (0.03) | 0.07 (0.03) | 0.22 |
| Max framewise displacement | 0.56 (0.62) | 0.54 (0.42) | 0.83 |
| Self-reported ethnicity/race | |||
| Asian | 1 | 2 | |
| Black or African American | 2 | 5 | |
| Hispanic/White | 13 | 12 | |
| Non-Hispanic/White | 18 | 16 | |
| Other/mixed | 7 | 6 | |
| Total OXTR risk alleles | 2.95 (1.05) | 3.26 (0.98) | 0.16 |
| Subjects (N) with ⩾ 1 risk allele | |||
| OXTR rs1042778 | 28 | 33 | |
| OXTR rs2254298 | 10 | 8 | |
| OXTR rs53576 | 15 | 23 | |
| OXTR rs237887 | 34 | 32 |
Abbreviations: ADI, Autism Diagnostic Interview; ADOS, Autism Diagnostic Observation Schedule; ASD, autism spectrum disorders; comm, communication; IQ, intelligence quotient; max, maximum; OXTR, oxytocin receptor gene; soc, social; TD, typically developing. Mean (s.d.). Subjects’ characteristics were matched across TD and ASD diagnostic groups.
MATERIALS AND METHODS
Participants
High-functioning youth with ASD (N = 56) and TD controls (N = 45) were recruited through the University of California, Los Angeles (UCLA’s) Center for Autism Research and Treatment or by flyers posted throughout the Los Angeles area. Participants with a history of claustrophobia, diagnosed neurological disorders, genetic conditions, structural brain abnormalities or metal implants were excluded from study participation. Study protocols were approved by the UCLA Institutional Review Board. Before assessment, informed consent and assent to participate in research were obtained from legal guardians and study participants. For ASD participants, inclusionary criteria included a prior diagnosis of autism confirmed at the UCLA Autism Evaluation Clinic with the Autism Diagnostic Observation Schedule – 2nd edition (ADOS-2),27 the Autism Diagnostic Interview – Revised (ADI-R),28 and best clinical judgment. Table 1 displays mean scores on social and communication subscales of the ADOS and ADI, as well as mean verbal, performance and full-scale IQ for ASD and TD participants, as assessed with the Wechsler Intelligence Scale for Children – 3rd edition (WISC)29 or the Wechsler Abbreviated Scale of Intelligence (WASI).30 Twenty-seven of the ASD children reported current use of psychotropic medications; 13 children reported use of a single medication, 14 children were on a combination of one or more medications (Supplementary Information).
Behavioral measures
Behavioral measures included the Social Responsiveness Scale (SRS)31 and the ADOS-2.27 The SRS is a parent-report questionnaire intended for use in children 4–18 years of age that provides a quantitative measure of social impairment. Autism severity scores were calculated for each of the ASD participants by converting ADOS-2 raw total scores into calibrated severity scores, which take into account chronological age and language level in assessing the severity of core autism features.32
Genotyping
DNA was extracted from saliva samples using standard protocols from the OraGene Collection Kit (DNA GenoTek, Ottawa, ON, Canada). The SNPs rs53576 and rs2254298 were genotyped at the UCLA Genotyping and Sequencing Core (http://genoseq.ucla.edu) using a 5′ nuclease assay to discriminate between the two alleles (Taqman SNP Genotyping Assay, Applied Biosystems, Foster City, CA, USA). Polymerase chain reactions were performed using 5-μl reaction volumes in 384-well plates with 25 ng of DNA and Taqman genotyping master mix from Applied Biosystems Inc. The standard protocol provided with the kit was followed. End point reads of fluorescence levels were obtained with an ABI 7900HT Sequence Detection System. Genome-wide SNP data, including rs237887 and rs1042778, were generated by the UCLA Neuroscience Genomics Core (https://www.semel.ucla.edu/ungc) using the Illumina Omni-1 or Omni-2.5-exome platforms according to standard manufacturer protocols. After quality filtering (<5% missing per person/per SNP, >1% minor allele frequency, Hardy–Weinberg equilibrium P>10−7), multi-dimensional scaling was performed in PLINK (http://pngu.mgh.harvard.edu/purcell/plink/)33 using the default settings with the HapMap 3 reference panel (http://hapmap.ncbi.nlm.nih.gov/).34
MRI data acquisition
Resting-state functional connectivity magnetic resonance imaging (MRI) data were collected on a Siemens 3T Trio whole-body scanner using a 16-channel phased-array head coil. For each subject, a scout localizing scan was first acquired for graphic prescription followed by a T2-weighted echo planar imaging volume (TR = 5000 ms, TE = 34 ms, 128 × 128 matrix size, 19.2 cm FoV, thirty-six 4mm axial slices, in plane voxel dimension = 1.50 × 1.50 mm), which was acquired co-planar to the functional volumes to ensure identical distortion characteristics. For the resting-state functional connectivity MRI data, a 6-min T2*-weighted functional scan was acquired while the subjects were asked to fixate on a black crosshair presented on a white screen (TR = 3000 ms, TE = 28 ms, 64 × 64 matrix size, 19.2 cm FoV, thirty-four 4 mm slices, in plane voxel dimension = 3.0 × 3.0 mm). To ensure that subjects were at ease during the scanning session, all participants participated in a mock scan before the date of their MRI.
Resting-state data processing
Neuroimaging data were analyzed using FMRIB’s Software Library (FSL, www.fmri.ox.ac.uk/fsl)35 and AFNI (Analysis of Functional NeuroImages).36 In brief, volumes were skull stripped and functional data were motion corrected to the average functional volume using FSL’s Motion Correction Linear Registration Tool (MCFLIRT);37 translations in the x, y and z dimensions were calculated from volume to volume, then averaged to create a measure of mean displacement. Subjects with >3 mm of motion from one volume to the next were excluded from further analyses (N = 7 ASD, N = 1 TD). Functional data were linearly registered to the high-resolution T2-weighted echo planar imaging volume (6 degrees of freedom (DOF)), then to the MNI152 2 mm standard brain (12 DOF) in light of empirical evidence indicating that the use of an adult brain template is appropriate for children aged 7 and above.38,39 Next, FSL’s Automatic Segmentation Tool was used to segment the high-resolution scans, creating subject-specific masks of gray matter, white matter and cerebral spinal fluid. The data were then band-pass filtered (0.1 Hz>t>0.01 Hz), smoothed (full width at half maximum 5 mm), and FSL’s fMRI Expert Analysis Tool (FEAT) was used to regress out nuisance variables including white matter, cerebral spinal fluid and global time-series. Volumes for which framewise displacement exceeded 0.5 mm and BOLD percent signal change from the prior volume exceeded 0.5% were removed (‘scrubbed’) from the data; one volume immediately proceeding and two volumes following the scrubbed volume were also removed.40 Volumes were removed for 13 ASD youth (mean volumes removed = 8.6, range = 4–19) and 13 TD youth (mean volumes removed = 9.1, range = 4–18); there was no difference between diagnostic groups in the number of removed volumes (P = 0.89). After scrubbing, participants with <5-min of resting-state functional connectivity MRI data were excluded from further analyses (N = 8 ASD, N = 3 TD), leaving a final sample of 41 youth with ASD and 41 TD youth. Power calculations were conducted with G*Power 3.1;41 according to previously published imaging-genetics effect sizes,23 our sample size is sufficient to detect significant effects at 80% power and a 0.05 significance level. All volume removal was completed before subsequent statistical analysis of the data. Finally, using an affine transformation, residuals from the motion scrubbed data were aligned to the T2-weighted high-resolution volume (6 DOF), then to the MNI one hundred fifty-two 2mm standard brain (12 DOF) with FMRIB’s Linear Image Registration Tool.
Resting-state data analysis
To examine functional connectivity of the NAcc, time series were extracted from the bilateral NAcc defined using the Harvard-Oxford Atlas at a threshold of 25% probability. Averaged time series from this region of interest were correlated with time series from every other voxel in the brain to generate maps of NAcc functional connectivity for each participant. Correlation maps were converted to z-statistic maps (Fisher’s r to z transform). Single-subject maps were then combined at the group level and compared using FEAT version 5.98, using separate estimates of variance for each diagnostic group. Whole-brain connectivity maps were compared between ASD and TD participants (Figure 1, Supplementary Table 1). For imaging-genetics analysis, a risk score was calculated for each subject by summing the number of risk alleles inherited across the four SNPs of interest (Supplementary Figure 1A). The group mean was then calculated across ASD and TD participants and used to de-mean single-subject risk scores. Demeaned values were used as covariates in higher-level FEAT analyses using FMRIB’s Local Analysis of Mixed Effects (FLAME). To identify brain areas with different OXTR modulation effects in the TD and ASD groups, the interaction effect was tested focusing on areas showing significant OXTR modulation in either group. All imaging results are presented at z>2.3, P<0.01, cluster corrected for multiple comparisons at P<0.05.
Figure 1.
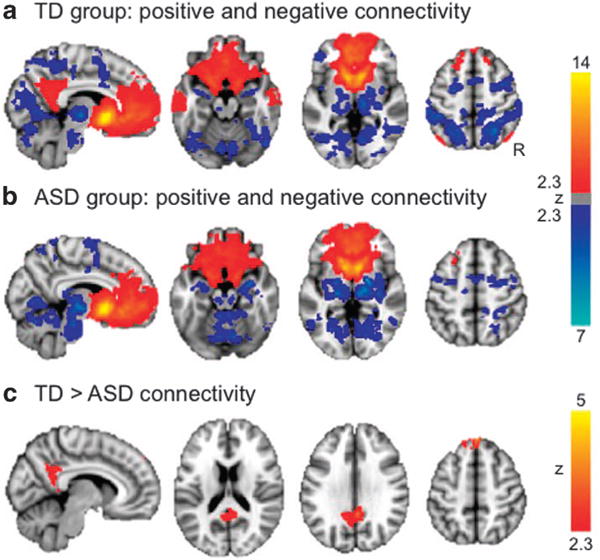
NAcc whole-brain connectivity. Warm colors indicate positive connectivity with the seed region; cool colors represent negative connectivity. (a) Connectivity in the TD group. (b) Connectivity in the ASD group. Results for (a and b) presented at z>2.3, P<0.01, cluster corrected for multiple comparisons at P<0.05. (c) Areas for which NAcc showed differential connectivity between diagnostic groups, z>2.3, P<0.01, cluster corrected for multiple comparisons at P<0.05. See also Supplementary Table 1. ASD, autism spectrum disorder; NAcc, nucleus accumbens; TD, typically developing.
RESULTS
Additive OXTR risk
The average number of risk alleles for all participants was 3.11 (s.d. = 1.02, range 1–6); neurotypical participants had an average of 2.95 risk alleles (s.d. = 1.05); participants with ASD had an average of 3.26 risk alleles (s.d. = 0.98; Supplementary Figures 1B and D). An independent samples t test showed there were no significant differences in the aggregate number of risk alleles between TD and ASD groups. Risk allele frequencies were equally distributed with skewness of 0.278 (s.e. = 0.266) and kurtosis of − 0.337 (s.e. = 0.526). To ensure that each SNP was inherited independently of the others, pairwise linkage disequilibrium (LD) was calculated in Haploview version 4.2;42 pairwise r2 in the data were <0.32 (Supplementary Figures 1C and E).
NAcc functional connectivity
As expected, the NAcc showed positive connectivity with other regions of the reward network including the bilateral caudate, putamen, anterior cingulate and frontal cortex, as well as negative connectivity with the thalamus, superior parietal and occipital brain areas (Figures 1a and b, Supplementary Table 1). Although overall similar patterns of positive and negative connectivity were found in both groups, direct between-group comparisons showed that youth with ASD had less connectivity between NAcc and superior frontal gyrus, as well as posterior cingulate/precuneus (a hub of the default mode network; Figure 1c, Supplementary Table 1).
Modulation by aggregate OXTR risk
In youth with ASD, carrying more OXTR risk alleles was associated with reduced NAcc connectivity with other nodes of the reward circuitry, specifically bilateral caudate, putamen and anterior cingulate gyrus (Figure 2a, Supplementary Table 2) – these results were not influenced by medication status (see Supplementary Information); no decreased connectivity was observed in TD youth as a function of higher aggregate risk score. In contrast, TD youth carrying more OXTR risk alleles showed increased NAcc connectivity with regions in the middle prefrontal cortex (Figure 2b, Supplementary Table 2); no increased connectivity was seen in ASD youth as a function of higher aggregate risk score. Next, to identify brain areas with significantly different OXTR modulation effects in the TD and ASD groups, the interaction effect was tested. Results confirmed that, as compared with their TD counterparts, ASD youth showed significantly reduced connectivity with striatal and mesolimbic regions, whereas TD participants showed significantly stronger NAcc connectivity with frontal cortex (Supplementary Figure 2).43 Iterative analyses of 3-SNP models confirmed that the observed genetic effects on NAcc functional connectivity were indeed additive across the 4 OXTR SNPs and that results were not disproportionately driven by any one SNP alone (Supplementary Information).
Figure 2.
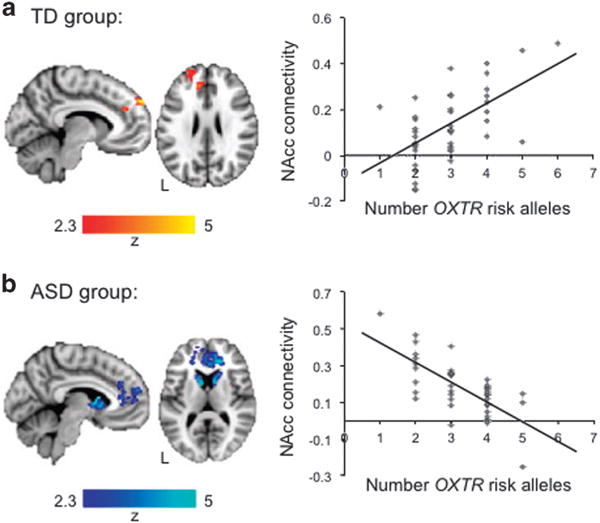
Effects of aggregate OXTR risk on functional connectivity (z>2.3, P<0.01, cluster corrected for multiple comparisons at P<0.05). (a) TD group: areas showing greater connectivity with the NAcc as a function of greater numbers of OXTR risk alleles. No increased functional connectivity with the NAcc as a function of OXTR risk alleles was observed in the ASD group (not shown). (b) ASD group: areas showing less connectivity with the NAcc as a function of greater OXTR aggregate risk. No decreased functional connectivity with the NAcc as a function of OXTR risk alleles was observed in the TD group (not shown). Scatterplots are shown for illustrative purposes and represent connectivity values extracted from the regions displayed at left as a function of number of OXTR risk alleles. See also Supplementary Table 2. ASD, autism spectrum disorder; NAcc, nucleus accumbens; OXTR, oxytocin receptor gene; TD, typically developing.
To evaluate whether imaging-genetics findings were influenced by ancestry, the first two components from multi-dimensional scaling of genome-wide data were controlled for in a correlation between OXTR aggregate risk score and connectivity values in regions modulated by risk-allele dosage. The observed correlations between aggregate OXTR-risk and NAcc connectivity in both ASD and TD groups remained significant (P<0.001).
Correlations with social responsiveness
Average connectivity values between the NAcc and regions modulated by the OXTR in TD and ASD groups were extracted and correlated with t-scores from subscales of the SRS.31 Although there were no significant findings in the ASD group, greater connectivity between the NAcc and frontal cortex in the TD group was correlated with better scores on the social cognition subscale of the SRS (r = − 0.35, P<0.05; Figure 3). To confirm these findings, we conducted a separate, independent analysis using SRS social cognition t-scores as a regressor of interest in a bottom–up analysis. Verifying our original finding, a cluster in the frontal pole reached statistical significance (peak coordinate –20, 46, 28, cluster size 63 voxels), indicating that more NAcc-frontal pole connectivity in the TD group is associated with better scores of social functioning. As each of the four OXTR SNPs used to create the aggregate risk score has independently been associated with ASD, we further hypothesized that inheritance of more risk-alleles would be related to greater ASD symptom severity. Indeed, in ASD participants, the correlation between OXTR risk-allele dosage and ADOS27 severity score was significant (r(40) = 0.28, P<0.05, one-tailed), such that individuals with greater numbers of OXTR risk alleles displayed more severe ASD symptomatology.
Figure 3.
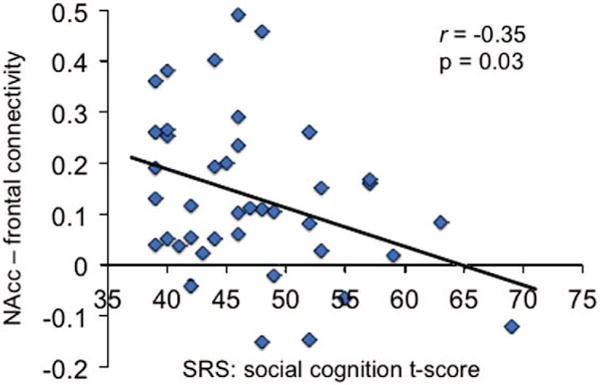
Behavioral correlation. Relationship between NAcc-frontal cortex connectivity in the TD group and SRS social cognition t-scores. NAcc, nucleus accumbens; SRS, Social Responsiveness Scale; TD, typically developing.
DISCUSSION
Here we show, for we believe the first time, that common ASD-associated genetic variants in the OXTR act additively to impact functional connectivity in the human brain. Specifically, we found that OXTR risk alleles coalesce to decrease connectivity between brain areas critical for reward processing in youth with ASD. These additive effects on brain circuits in individuals with ASD are consistent with evidence suggesting that ASDs follow a polygenic pattern of inheritance, with genetic variants at many loci contributing to expression of the ASD phenotype.1 In our ASD sample, greater OXTR risk-allele dosage was associated with more severe ASD symptomatology as indexed by the ADOS, demonstrating that cumulative risk on this gene does exert significant negative effects on social functioning in individuals with ASD. The oxytocin receptor is a g-protein-coupled receptor that, through a cascade of events, affects intracellular calcium levels and neural excitability.44 Thus, alterations in neural excitability in the NAcc – the hub of the reward circuit – may explain our findings of reduced functional connectivity, as well as previous reports of hypoactivity during reward processing in ASD.15,16 Furthermore, we found that in both youth with ASD and neurotypical controls, greater genetic risk in the OXTR modulates NAcc functional connectivity, although in different ways. Specifically, greater OXTR risk in ASD children was associated with decreased connectivity in the striatum, a set of brain regions critical for reward processing and implicit learning; conversely, greater OXTR risk in neurotypical children was associated with increased connectivity with the frontal pole, an area implicated in the ability to understand the mental states of the self and others.
Remarkably, our observations of reduced connectivity as a function of increased genetic risk were limited to individuals with ASD; significant interaction effects confirmed that, compared with their TD counterparts, ASD youth showed significantly reduced NAcc connectivity with subcortical brain regions implicated in reward processing, whereas TD youth showed stronger NAcc connectivity with frontal regions implicated in mentalizing processes.43 This finding of increased connectivity in TD children could reflect a compensatory mechanism in the face of increased genetic risk. Consistent with this interpretation, we found that greater NAcc-frontal pole connectivity in TD youth was related to better social cognition measured with the SRS.31 These findings are in agreement with prior imaging-genetics work showing that neurotypical individuals who are carriers of ASD-risk variants show alterations in brain function and structure.45,46 Our results in neurotypical youth highlight possible compensatory mechanisms, which may lead to improved social cognition despite increased OXTR risk-allele dosage. Importantly, there is also evidence that genetic variants may have enhanced effects (that is, increased penetrance) on disease-related brain circuits in individuals who have a diagnosis of ASD47 – an effect likely due to the presence of other genetic (that is, epistatic) and environmental susceptibilities. Our data indicate that additive genetic effects in the OXTR have more pronounced effects on connectivity with reward-related brain circuits in individuals who express the ASD phenotype. More generally, these findings indicate that additive genetic vulnerability in biological pathways underlying reward processing in the human brain may be one mechanism by which neural vulnerability leads to atypical social behavior in ASD.
Although the present study focused on the effects of multiple SNPs in a single gene, it will be important for future research to examine risk factors across multiple genes implicated in autism, which may modulate brain activity and connectivity in other neural circuits known to be compromised in ASD. Our study focused on a small set of ASD-associated OXTR SNPs that demonstrated a lack of LD with one another to assure statistical independence of each SNP’s inheritance. Additional variance in neural endophenotypes may be explained by using more complex statistical models to account for the degree of LD between loci and thus examine the additive effects of ASD-associated SNPs in close proximity to one another. Although there is some evidence that several OXTR SNPs affect expression of the oxytocin receptor in the human brain (that is, they are expressed quantitative trait loci; http://www.braineac.org), these effects are not genome-wide significant, thus limiting our ability to relate our results to variation in NAcc oxytocin receptor expression. Nevertheless, our findings show that functional connectivity in the reward network varies significantly with OXTR risk-allele dosage and provide a model for integrating genetic risk across multiple loci with neuroimaging data to further elucidate mechanisms of vulnerability and resilience to neurocognitive disorders.
Given recent interest in the use of intranasal oxytocin as a treatment option for individuals with ASD,48 our results also have broad clinical relevance. Several studies have shown that intranasal oxytocin administration affects brain activity during social-emotional processing in neurotypical individuals49 and in individuals with ASD;50–53 further, oxytocin administration has been shown to rescue social behaviors in a mouse model of ASD.54 Importantly, the effects of intranasal administration on brain activity in humans may be moderated by genotypic variation in the OXTR55 and other genes in the same pathway. For instance, during a task designed to elicit social cooperation, neurotypical male and female carriers of the OXTR rs53576 GG genotype show different patterns of ventral striatal activity in response to oxytocin administration, a sex-specific effect not seen in carriers of other rs53576 genotypes.56 Similarly, when exposed to intranasal oxytocin, individuals homozygous for the ASD-associated allele of rs3796863 in the CD38 gene (involved in endogenous oxytocin secretion) show increased brain activity in the fusiform gyrus during a face processing task, an effect again not seen in individuals carrying non-risk alleles.57 Together, these studies suggest that aggregate genetic variability may be an important biomarker to consider when determining which individuals would optimally benefit from oxytocin treatment. The identification of subgroups, within the heterogeneous ASD population, is a prerequisite step to implement targeted interventions; our findings are relevant to this goal as they indicate that stratification by risk-allele dosage may reveal distinct neural endophenotypes and thus help parse the considerable heterogeneity observed in ASD.
Overall, our results expand on other lines of evidence, from both animal models and human studies, indicating that genetic variation in the OXTR is related to reward system functioning. Importantly, as the observed additive genetic effects differed in youth with and without ASD, this work underscores the benefits of examining cumulative genetic risk to elucidate neural pathways conferring increased vulnerability, as well as resilience, in carriers of disease-associated risk alleles. Our findings of additive genetic effects on neural endophenotypes indicate that future research should embrace complex genetic variability by examining aggregate risk both within disease-associated genes and across genes in disease-associated biological pathways.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Child Health and Human Development grant P50 HD055784 (to SYB), National Institute of Mental Health grant RO1 MH100028 (to SYB), National Institute of Mental Health grant T32 MH073526-08 (to LMH), National Research Service Awards F32 MH105167-01 (to SAG) and F31 DA038578-01A1 (to LES). We are grateful for the generous support from the Brain Mapping Medical Research Organization, Brain Mapping Support Foundation, Pierson-Lovelace Foundation, The Ahmanson Foundation, Capital Group Companies Charitable Foundation, William M. and Linda R. Dietel Philanthropic Fund, and Northstar Fund. Research reported in this publication was also partially supported by the National Center for Research Resources and by the Office of the Director of the National Institutes of Health under award numbers C06RR012169, C06RR015431 and S10OD011939. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp).
References
- 1.Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey JA, et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huguet G, Ey E, Bourgeron T. The genetic landscapes of autism spectrum disorders. Annu Rev Genomics Hum Genet. 2013;14:191–213. doi: 10.1146/annurev-genom-091212-153431. [DOI] [PubMed] [Google Scholar]
- 4.LoParo D, Waldman ID. The oxytocin receptor gene (OXTR) is associated with autism spectrum disorder: a meta-analysis. Mol Psychiatry. 2014;20:640–646. doi: 10.1038/mp.2014.77. [DOI] [PubMed] [Google Scholar]
- 5.Yrigollen CM, Han SS, Kochetkova A, Babitz T, Chang JT, Volkmar FR, et al. Genes controlling affiliative behavior as candidate genes for autism. Biol Psychiatry. 2008;63:911–916. doi: 10.1016/j.biopsych.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wermter AK, Kamp-Becker I, Hesse P, Schulte-Körne G, Strauch K, Remschmidt H. Evidence for the involvement of genetic variation in the oxytocin receptor gene (OXTR) in the etiology of autistic disorders on high-functioning level. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:629–639. doi: 10.1002/ajmg.b.31032. [DOI] [PubMed] [Google Scholar]
- 7.Campbell DB, Datta D, Jones ST, Lee EB, Sutcliffe JS, Hammock EAD, et al. Association of oxytocin receptor gene (OXTR) gene variants with multiple phenotype domains of autism spectrum disorder. J Neurodev Disord. 2011;3:101–112. doi: 10.1007/s11689-010-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Insel TR, Shapiro LE. Oxytocin receptor distribution reflects social organization in monogamous and polygamous voles. Proc Natl Acad Sci USA. 1992;89:5981–5985. doi: 10.1073/pnas.89.13.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett CE, Arambula SE, Young LJ. The oxytocin system promotes resilience to the effects of neonatal isolation on adult social attachment in female prairie voles. Transl Psychiatry. 2015;5:e606. doi: 10.1038/tp.2015.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King LB, Walum H, Inoue K, Eyrich NW, Young LJ. Variation in the oxytocin receptor gene predicts brain region-specific expression and social attachment. Biol Psychiatry. 2016;80:160–169. doi: 10.1016/j.biopsych.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young LJ, Lim M, Gingrich B, Insel TR. Cellular mechanisms of social attachment. Horm Behav. 2001;40:133–148. doi: 10.1006/hbeh.2001.1691. [DOI] [PubMed] [Google Scholar]
- 12.Keebaugh AC, Barrett CE, Laprairie JL, Jenkins JJ, Young LJ. RNAi knockdown of oxytocin receptor in the nucleus accumbens inhibit social attachment and parental care in monogamous female prairie voles. Soc Neurosci. 2015;7:1–10. doi: 10.1080/17470919.2015.1040893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dölen G, Darvishzadeh A, Huang KW, Malenka RC. Social reward requires coordinated activity of nucleus accumbens oxytocin and serotonin. Nature. 2013;501:179–184. doi: 10.1038/nature12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitz N, Rubia K, van Amelsvoort T, Daly E, Smith A, Murphy DG. Neural correlates of reward in autism. Br J Psychiatry. 2008;192:19–24. doi: 10.1192/bjp.bp.107.036921. [DOI] [PubMed] [Google Scholar]
- 15.Scott-Van Zeeland AA, Dapretto M, Ghahremani DG, Poldrack RA, Bookheimer SY. Reward processing in autism. Autism Res. 2010;3:53–67. doi: 10.1002/aur.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dichter GS, Richey JA, Rittenberg AM, Sabatino A, Bodfish JW. Reward circuitry function in autism during face anticipation and outcomes. J Autism Dev Disord. 2012;42:147–160. doi: 10.1007/s10803-011-1221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohls G, Schulte-Rüther M, Nehrkorn B, Müller K, Fink GR, Kamp-Becker I, et al. Reward system dysfunction in autism spectrum disorders. Soc Cogn Affect Neurosci. 2013;8:565–572. doi: 10.1093/scan/nss033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delmonte S, Balsters JH, McGrath J, Fitzgerald J, Brennan S, Fagan AJ, et al. Social and monetary reward processing in autism spectrum disorders. Mol Autism. 2012;3:7. doi: 10.1186/2040-2392-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson G, Toth K, Abbott R, Osterling J, Munson J, Estes A, et al. Early social attention impairments in autism: social orienting, joint attention, and attention to distress. Dev Psychol. 2004;7:340–359. doi: 10.1037/0012-1649.40.2.271. [DOI] [PubMed] [Google Scholar]
- 20.Chevallier C, Kohls G, Troiani V, Brodkin ES, Schultz RT. The social motivation theory of autism. Trends Cogn Sci. 2012;16:231–239. doi: 10.1016/j.tics.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tost H, Kolachana B, Hakimi S, Lemaitre H, Verchinski BA, Mattay VS, et al. A common allele in the oxytocin receptor gene (OXTR) impacts prosocial temperament and human hypothalamic-limbic structure and function. Proc Natl Acad Sci USA. 2010;107:13936–13941. doi: 10.1073/pnas.1003296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito Y, Suga M, Tochigi M, Abe O, Yahata N, Kawakubo Y, et al. Neural correlate of autistic-like traits and a common allele in the oxytocin receptor gene. Soc Cogn Affect Neurosci. 2014;9:1443–1450. doi: 10.1093/scan/nst136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Damiano CR, Aloi J, Dunlap K, Burrus CJ, Mosner MG, Kozink RV, et al. Association between the oxytocin receptor (OXTR) gene and mesolimbic responses to rewards. Mol Autism. 2014;5:7. doi: 10.1186/2040-2392-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu S, Jia M, Ruan Y, Liu J, Guo Y, Shuang M, et al. Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biol Psychiatry. 2005;58:74–77. doi: 10.1016/j.biopsych.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Lerer E, Levi S, Salomon S, Darvasi A, Yirmiya N, Ebstein RP. Association between the oxytocin receptor (OXTR) gene and autism: relationship to Vineland Adaptive Behavior Scales and cognition. Mol Psychiatry. 2008;10:980–988. doi: 10.1038/sj.mp.4002087. [DOI] [PubMed] [Google Scholar]
- 26.Di Napoli A, Warrier V, Baron-Cohen S, Chakrabarti B. Genetic variation in the oxytocin receptor (OXTR) gene is associated with Asperger Syndrome. Mol Autism. 2014;5:48. doi: 10.1186/2040-2392-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lord C, Rutter M, DiLavore P, Risi S, Gotham K, Bishop S. Autism Diagnostic Observation Schedule. 2nd. Western Psychological Services; Torrance, CA, USA: 2012. [Google Scholar]
- 28.Lord C, Rutter M, Le Couteur A. Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 29.Wechsler D. The Wechsler Intelligence Scale For Children. The Psychological Corporation; San Antonio, TX, USA: 1991. [Google Scholar]
- 30.Wechsler D. Wechsler Abbreviated Scale of Intelligence. The Psychological Corporation, Harcourt Brace & Company; New York, NY, USA: 1999. [Google Scholar]
- 31.Constantino JN, Gruber CP. Social Responsiveness Scale (SRS) Manual. Western Psychological Services; Los Angeles, CA, USA: 2005. [Google Scholar]
- 32.Gotham K, Pickles A, Lord C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J Autism Dev Disord. 2009;39:693–705. doi: 10.1007/s10803-008-0674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The International HapMap Consortium. The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 35.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 36.Cox RW. AFNI software for analysis and visualization of functional magnetic resonance images. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 37.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brains. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 38.Burgund ED, Kang HC, Kelly JE, Buckner RL, Snyder AZ, Petersen SE, et al. The feasibility of a common stereotactic space for children and adults in fMRI studies of development. Neuroimage. 2002;17:184–200. doi: 10.1006/nimg.2002.1174. [DOI] [PubMed] [Google Scholar]
- 39.Kang HC, Burgund ED, Lugar HM, Petersen SE, Schlaggar BL. Comparison of functional activation foci in children and adults using a common stereotactic space. Neuroimage. 2003;19:16–28. doi: 10.1016/s1053-8119(03)00038-7. [DOI] [PubMed] [Google Scholar]
- 40.Power JD, Barnes KA, Snyder AZ, Schlaggar B, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faul F, Erdfelder E, Buchner A, Lang A-G. Statistical power analyses using G*Power 3,1: tests for correlation and regression analyses. Behav Res Methods. 2009;41:1149–1160. doi: 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
- 42.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 43.Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 2011;8:665–670. doi: 10.1038/nmeth.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gimpl G, Fahrenholz F. The oxytocin receptor system: structure, function, and regulation. Physiol Rev. 2001;82:629–683. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]
- 45.Dennis EL, Jahanshad N, Rudie JD, Brown JA, Johnson K, McMahon KL, et al. Altered structural brain connectivity in healthy carriers of the autism risk gene, CNTNAP2. Brain Connect. 2011;1:447–459. doi: 10.1089/brain.2011.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whalley HC, O’Connell G, Sussmann JE, Peel A, Stanfield AC, Hayiou-Thomas ME, et al. Genetic variation in CNTNAP2 alters brain function during linguistic processing in healthy individuals. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:941–948. doi: 10.1002/ajmg.b.31241. [DOI] [PubMed] [Google Scholar]
- 47.Rudie JD, Hernandez LM, Brown JA, Beck-Pancer D, Colich NL, Gorrindo P, et al. Autism-associated promoter variant in MET impacts functional and structural brain networks. Neuron. 2012;75:904–915. doi: 10.1016/j.neuron.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young LJ, Barrett C. Can oxytocin treat autism? Science. 2015;347:825–826. doi: 10.1126/science.aaa8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zink C, Meyer-Lindenberg A. Human neuroimaging of oxytocin and vasopressin in social cognition. Horm Behav. 2012;61:400–409. doi: 10.1016/j.yhbeh.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Domes G, Heinrichs M, Kumbier E, Grossmann A, Hauenstein K, Herpertz SC. Effects of intranasal oxytocin on the neural basis of face processing in autism spectrum disorder. Biol Psychiatry. 2013;74:164–171. doi: 10.1016/j.biopsych.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Gordon I, Vander Wyk BC, Bennett RH, Cordeaux C, Lucas MV, Eilbott JA, et al. Oxytocin enhances brain function in children with autism. Proc Natl Acad Sci USA. 2013;110:20953–20958. doi: 10.1073/pnas.1312857110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Domes G, Kumbier E, Heinrichs M, Herpertz SC. Oxytocin promotes facial emotion recognition and amygdala reactivity in adults with Asperger syndrome. Neuropsychopharmacology. 2014;39:698–706. doi: 10.1038/npp.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe T, Kuroda M, Kuwabara H, Aoki Y, Iwashiro N, Tatsunobu N, et al. Clinical and neural effects of six-week administration of oxytocin on core symptoms of autism. Brain. 2015;138:3400–3412. doi: 10.1093/brain/awv249. [DOI] [PubMed] [Google Scholar]
- 54.Peñagarikano O, Lázaro MT, Lu XH, Gordon A, Dong H, Lam HA, et al. Exogenous and evoked oxytocin restores social behavior in the Cntnap2 mouse model of autism. Sci Transl Med. 2015;7:271ra8. doi: 10.1126/scitranslmed.3010257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen FS, Kumsta R, Dvorak F, Domes G, Yim OS, Ebstein R, et al. Genetic modulation of oxytocin sensitivity: a pharmacogenetic approach. Transl Psychiatry. 2015;5:e664. doi: 10.1038/tp.2015.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng C, Lori A, Waldman ID, Binder EB, Haroon E, Rilling JKP. A common oxytocin receptor gene (OXTR) polymorphism modulates intranasal oxytocin effects on the neural response to social cooperation in humans. Genes Brain Behav. 2015;14:516–525. doi: 10.1111/gbb.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauer C, Montag C, Wörner C, Kirsch P, Reuter M. Effects of a common variant in the CD38 gene on social processing in an oxytocin challenge study: possible links to autism. Neuropsychopharmacology. 2012;37:1474–1482. doi: 10.1038/npp.2011.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


