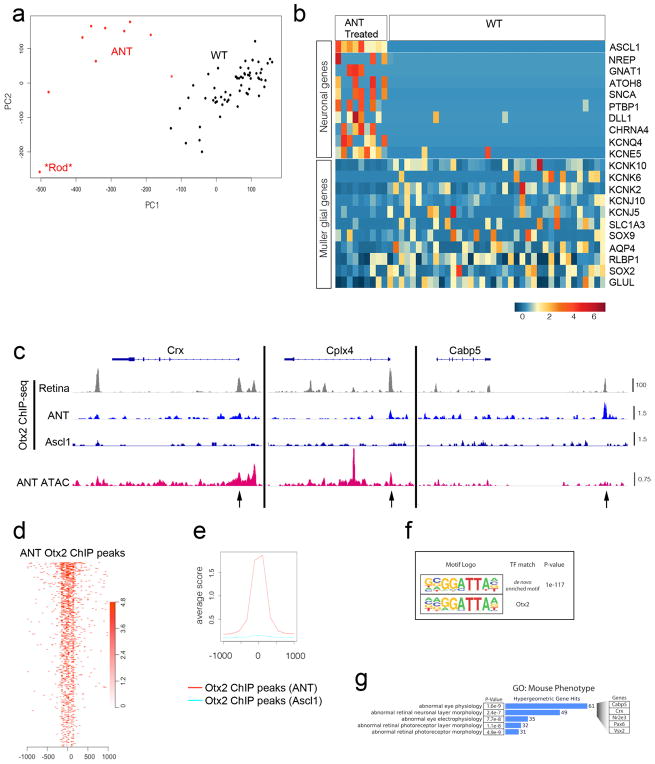
Extended Data Figure 9. FACS-purified ANT-treated cells show increased neuronal gene expression and Otx2 binding.
a, Principal component analysis from Fluidigm single-cell mRNA-seq showing 48 wild-type MG (black), 9 ANT-treated cells (red), and 1 cell that was probably a contaminating rod photoreceptor (asterisk). The ANT-treated cells formed a separate cluster from the wild-type MG. b, Heat map showing ANT-treated MG-derived cells and wild-type MG. Each column is an individual cell, with example neural and glial genes as rows. Reprogramming with ANT leads to downregulation of glial genes and an increase in neural genes. Scale is log2(CPM). ANT-treated retinas were collected 46 days post-TSA. c, IGV browser tracks at bipolar genes Crx, Cplx4 and Cabp5 showing: Otx2 ChIP–seq from whole adult mouse retina15 (grey); Otx2 ChIP–seq from FACS-purified ANT-treated and Ascl1-only cells (blue); and DNA accessibility from ATAC–seq of FACS-purified ANT-treated cells (red). Scale for all tracks shown to the right as reads per million. Black arrows mark open chromatin in promoter regions that are bound by Otx2 in ANT-treated but not control Ascl1 cells, and are appropriate regions of binding based on a whole-retina Otx2 ChIP–seq dataset. d, e, Otx2 ChIP–seq peaks from ANT-treated cells show central enrichment at Otx2 ChIP–seq sites from whole adult retina15, but this is not present in the Otx2 ChIP–seq from MG that expressed Ascl1 but received none of the other treatments (TSA, NMDA). f, Top-scoring transcription factor binding motif enrichment (HOMER software suite) from ANT-treated Otx2 ChIP–seq peak calls along with the closest matching transcription factor motif (TF match) and P value for de novo motif enrichment. g, Gene Ontology enrichment for category ‘Mouse Phenotype’ of peaks from d, showing P value for term enrichment, the number of associated genes (blue bars), and example genes from these enriched terms (expanded at right). ChIP–seq was performed on 490,430 and 692,271 pooled cells from 17 ANT-treated Rlbp1-Ascl1 and 15 untreated Rlbp1-Ascl1 mice, respectively. Retinas were collected 9–18 days post-TSA.