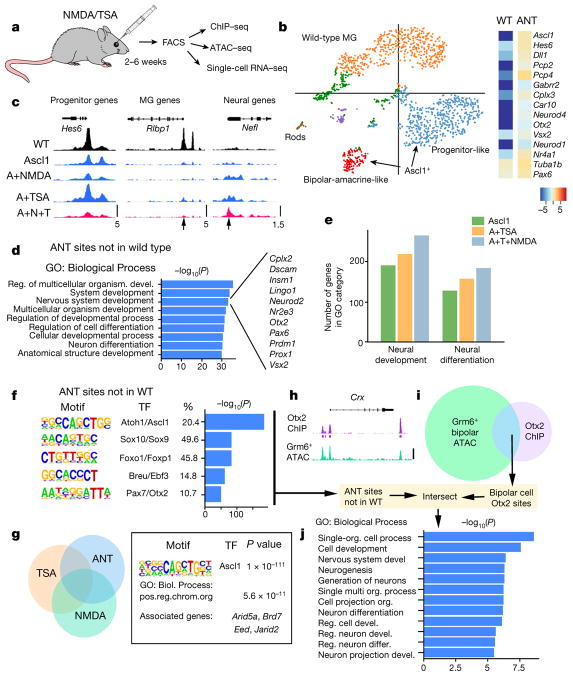
Figure 4. ANT treatment enables epigenetic changes and expression of neuronal genes in MG.
a, FACS-purified MG-derived neurons subjected to genomic analyses. b, k-means clustering of single-cell RNA-seq from wild-type (WT) and ANT-treated cells shown as a t-SNE plot. Heat map showing average expression of neural genes for all Ascl1+ cells in the ANT condition (19 days post-TSA) compared with wild type. c, ATAC–seq-normalized reads shown for sample progenitor (Hes6), MG (Rlbp1) and neural (Nefl) genes. DNA accessibility changes with Ascl1 expression (arrows). Scale at the bottom track for each set (reads per million, RPM). ANT-treated mice 15 days post-TSA. d, Gene Ontology enrichment for regions of increased DNA accessibility in ANT-treated cells compared with wild type. Examples of the ‘Nervous system development’ category are shown to the right. e, The number of genes in enriched GO categories from MG expressing Ascl1, Ascl1 and TSA, and from ANT-treated retinas. f, Top 5 transcription factor motifs in peaks of ANT, but not wild type; percentage of peaks with motif and enrichment score. g, ATAC–seq peaks for ANT versus TSA and NMDA were compared; ANT-specific peaks analysed for motif enrichment and Gene Ontology. Two of the top ten GO enriched Biological Processes were chromatin-remodelling genes. h, i, Otx2 ChIP–seq peaks from total retina15, was compared with ATAC–seq from Grm6+ FACS-purified bipolar cells and the overlapping (bipolar-specific Otx2 peaks) regions intersected with the ANT peaks that were not present in the wild type (scale bar, 100 counts per million reads (CPM) for Otx2-ChIP and 10 CPM for ATAC–seq). j, Gene Ontology enrichment for Biological Process of peaks that overlap with ANT and bipolar cell Otx2 sites showing P value for term enrichment for neural genes.