Abstract
Herbal drugs, on which 80% of the world's population rely, are relatively safe over conventional drugs. Conventional drugs are costly, have serious side effects and hence over the past few decades researchers have focused on drug discovery from herbal medicines or botanical sources. The majority of new herbal drugs have been generated from secondary metabolites (alkaloids, terpenoids and phenolic compounds) of plant metabolism. Till date, only a small fraction of the vast diversity of plant metabolism has been explored for the production of new medicines and other products. The emergence of new herbal genomics research, medicinal plant genomics consortium, together with advances in other omics information may help for the speedy discovery of previously unknown metabolic pathways and enzymes. This review highlights the importance of genomics research in the discovery of some previously unknown enzymes/pathways which may make significant contributions in plant metabolic biology and may be used for the future discovery of many new pharmaceutical agents.
Keywords: Herbal, Genomics, Metabolite, Drug
Highlights
-
•
New herbal drugs generated from secondary metabolites of plant metabolism.
-
•
Genome research can find gene clusters and gene duplication events responsible for specialized metabolism in plants.
-
•
Genome and other omic research helps to find genes to metabolite link.
-
•
This tool could be used for discovery of new pharmaceutical agents.
1. Introduction (why herbal genomics?)
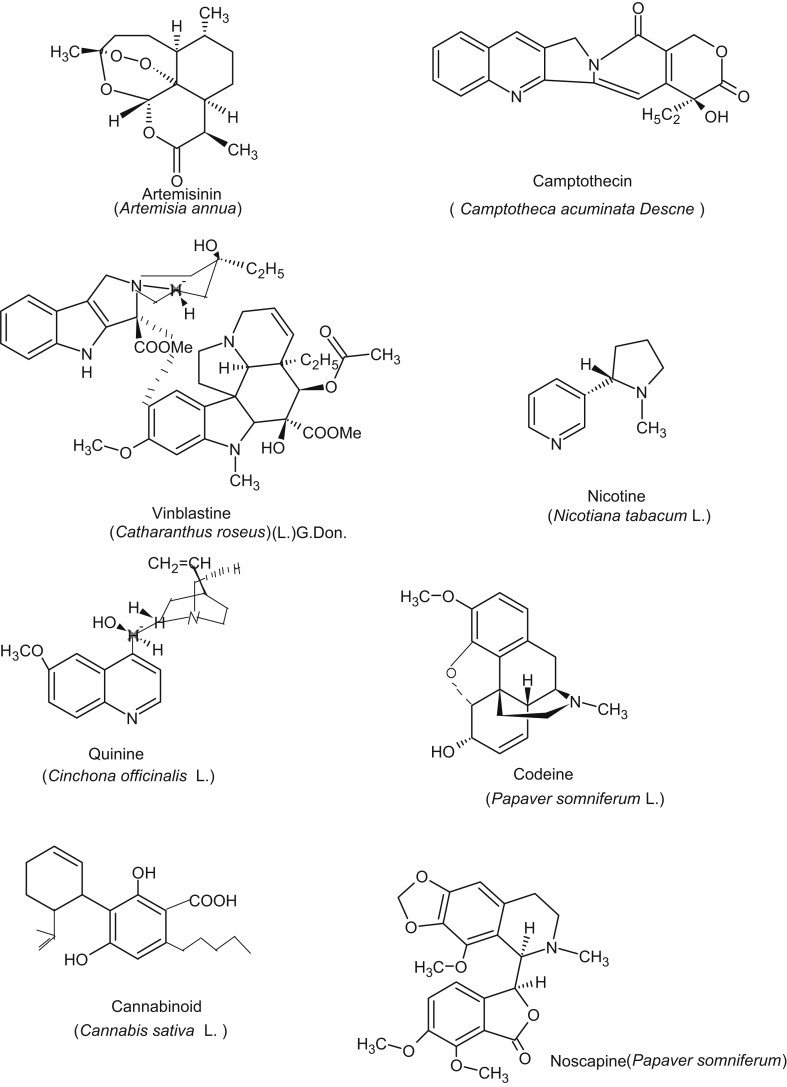
Traditional herbal medicines, basically plant-based therapies have been used for more than 5000 years [1]. At the time of invention of medicinal properties of herbs, people could not think of scientific evidence, philosophical and experimental basis, molecules responsible for medicinal value and of course the currently emerging herbal genomics. Constant and renewed public interest on alternative and complementary medicine lies mainly due to high cost of new drugs, increased side effects, microbial resistance and lack of curative treatment for several chronic diseases. Countries like India, China, Korea and Japan are now taking lead role and continuously investing in research on evidence-based traditional medicines and scientific validation of fundamental principles. China, the traditional medicine giant has successfully promoted its own therapies over the globe with a science-based approach and evidenced by the rapid increase in number of licensed Chinese medicine providers in the United states [2]. India is also gearing up, and there has been a steep rise in the global acceptance of Ayurveda, the traditional Indian medicine. The World Health Organization has listed 21,000 medicinal plants, among which 2500 species are in India and now India is known as the largest producer of medicinal herbs [3]. Collectively, global data shows that 80% of world's population, rely primarily on ethnobotanical remedies and plant drugs, e.g. antineoplastic: camptothecin, Taxol, antimalarial: artemisinin, quinine, antigout: colchicine, analgesic: codeine, morphine, cardiac depressant: quinidine, antidiabetic: allicin, and for brain functions: caffeine, nicotine are the well known curative agents (Table 1, Fig. 5, modified from [4], [5]).
Table 1.
Recognized drugs from medicinal plants and their actions against various human diseases.
| Drugs | Medicinal plants | Actions against |
|---|---|---|
| vinblastine | Catharanthus roseus | Cancer |
| vincristine | Catharanthus roseus | |
| camptothecin | Camptotheca acuminata | |
| taxol | Taxus baccata | |
| podophyllotoxin | Podophyllum peltatum | |
| artemisinin | Artemisia annua | Malaria |
| quinine | Cinchona ledgeriana | |
| quinoline | Cinchona ledgeriana | |
| digoxin | Digitalis purpurea | Cardiac disorder |
| digitoxin | Digitalis purpurea | |
| quinidine | Cinchona ledgeriana | |
| codeine | Papaver somniferum | Analgesic |
| morphine | Papaver somniferum | |
| allicin | Allium sativum | Diabetes |
| SMCS (S-methyl cysteine sulfoxide) | Allium cepa | |
| nicotine | Nicotiana tabaccum | Brain disorder |
| caffeine | Coffea canephora | |
| diosgenin | Dioscorea Mexicana | sexual problems |
| stigmasterol | Glycine max | |
| cannabinoids | Cannabis sativa | psycho-disorder |
| tubocurarine | Chondodendron tomentosum | muscle disorder |
| atropine | Atropa belladonna | nervous system |
| hyoscyamine | Hysocyamus niger | |
Fig. 5.
Some important structures from medicinal plants and the names of plants that produce them.
Although, plant derived natural products remain rich resources for drug development and have had a profound and lasting impact on human health with almost 100 plant-derived compounds in clinical trials as of late 2007, the clinical potential of these compounds is often curtailed due to low production levels in plant species or due to loss of source for extinction [6]. For example, use of the blockbuster drug Taxol almost stopped in the early 1990's because the primary source, yew tree bark, could not be used as a sustainable source of the drug. A taxol precursor and a semi-synthetic protocol could have been the ideal way to convert it into the active drug. More generalized solutions, such as metabolic engineering of effective plant and microbial production platforms, are urgently needed to ensure that the plant-derived compounds having enormous structural diversity and biological activities enter the clinical pipeline and find widespread use in medicine.
Considering the vast chemical biodiversity of the plant world, and widespread medicinal values of plant derived natural products, researchers are now focusing on the poorly understood areas of herbs such as genetic background, the agricultural traits, and the medicinal quality. With rapid advances in high throughput sequencing technologies and greatly reduced costs, a new discipline called “herbal genomics” is now emerging. Systematic analysis of medicinal herbs genes' functions are necessary and achievable through sequencing, assembling and annotating their genomes. There have been only a few well assembled herbal genomes released to date, partly because of their complexity. Genomic information, together with transcriptomic, proteomic and metabolomic data, can therefore be used to predict the secondary metabolic pathways of herbs. Moreover, functional herbal genomics can contribute to model herb research platforms, geoherbal research, and herbal synthetic biology, all of which are important for securing the sourcing of the medicinal plants and their active compounds in the future.
2. Genomes and metabolic activities/pathways of herbs
Plants synthesize an abundance of metabolites that can be exploited for pharmacological purposes. Till today, only a small fraction of the immense diversity of plant metabolism has been explored for the production of new medicines and other products important to human well-being. All plants synthesize basic metabolites through primary metabolism needed for their survival but different taxa produce distinct metabolites through secondary or specialized metabolism that are specialized for some specific reactions. As stationary autotrophs, plants have to cope with a number of challenges such as local fluctuations of the simple nutrients they require to synthesize their foods, coexistence of herbivores and pathogens in their immediate environment. Plants have therefore evolved secondary biochemical pathways that allow them to synthesize a diverse array of organic compounds to counterattack to specific environmental stimuli [7], [8]. The palette of secondary metabolites are subdivided into a number of distinct groups, on the basis of their chemical structure and synthetic pathways and those groups are the alkaloids, terpenoids and the phenolic compounds. With a view to understand the full metabolic potential/pathways of the plant/herbs, the whole nuclear and chloroplast genomes needed to be sequenced. In this regard, Chen et al. [44], [45] initiated a project, “Herb Genome Programme” for the genome sequencing of various medicinal plants and post genomic functional analysis of various secondary metabolite biosynthetic pathways. Unfortunately, there have been only a few well assembled herbal genomes released to date, partly because of their complexity. The genomes of some commonly used herbs such as Ganoderma lucidum, Salvia miltiorrihiza and Catharanthus roseus have already been sequenced and they emerged as valuable models for studying the genetics and metabolic activities of herbs [1], [9], [10], [50]. These species have been shown to synthesize active pharmaceutical components, including triterpenes, diterpene quinone and indole alkaloids. Analysis of a draft genome sequence of C. roseus provided evidence for partial clustering of genes for the biosynthesis of the monoterpene indole alkaloids vinblastine and vincristine. With the help of bacterial artificial chromosome (BAC) sequencing, Kellner et al. [50] showed seven small clusters each of two to three genes that contained genes encoding enzymes for vinblastine/vincristine biosynthesis pathway and other genes for other pathway. In addition to those, the whole genome of medicinal plant, Z. jujuba [46], [51], A.Indica(Neem) [47], and chloroplast genomes of P. cablin [48] and S. miltiorrihiza [49]have been sequenced successfully. Z. jujuba has got significant medicinal value and its fruits are rich in vitamin C and sugar, and the plant contains various therapeutically important alkaloids, flavonoids and phenolics. De novo assembly of its complex genome and transcriptomics data established that l-galactose pathway is the major synthesis pathway for vitamin C and consistently higher expression of the genes for the enzymes, GDP-d-mannose 3,5 epimerase, and GDP-l-galactose phosphorylase contributes for sugar metabolism. A. Indica (Neem) is important for its huge medicinal value and bioactivities against malaria, diabetes and tumor. The genomes and transcriptomes analysis of neem shows that its genome is AT-rich, bears little repetitive DNA elements and comprises about 20,000 genes. Comparative transcript expression analysis showed either exclusive or enhanced expression of known genes involved in neem terpenoid biosynthesis. Draft sequence of whole genome of S. miltiorrihiza by Xu et al. [52]. shows that plant genome size is ∼600 MB and contains 30,478 protein coding genes, and 1620 genes for transcription factors, and several of these transcription factors revealed to be involved in the biosynthesis of tanshinone and phenolic acids used in several cardiovascular, cerebrovascular and hyperlipidemia disease. While, chloroplast genome of this plant is 151,328 bp in length and it contains 114 unique genes including 80-protein coding genes, 30-tRNA genes and four rRNA genes [49]. The chloroplast genome sequences will facilitate population, phylogenetic and genetic engineering studies of this medicinal plant. Pogostemon cablin, an important herb in the Lamiaceae family contains more than 40 major components, including flavonoids, terpenoids, alkaloids and phenylpropanoid glycosides. It has got wide range of applications including medicinal effects like anti-inflammatory, antidepressant activity. The entire chloroplast genome with 38.24% GC content, is 152,460 bp in length and contains 127 genes of which 107 genes are single-copy, including 79 protein –coding genes, four rRNA genes, and 24 tRNA genes. Phylogenetic analysis reveals that P. cablin diverged from the Sentellarioideae clade about 29.45 million year ago. Complete sequences and annotation of P. cablin genome will help in the understanding of population, phylogenetic and genetic engineering research [48]. Ocimum sanctum L. is an important sacred medicinal plant of India, whose whole nuclear and chloroplast genomes recently been sequenced [11]. The genome sequence and annotation of O. sanctum and another species O. basilicum indicates higher expression of phenylpropanoid/terpenoid pathway genes, expression of several cytochrome P450s and transcription factor families and the information thus provides new insights for mining biosynthetic pathways for important metabolites of medicinal nature in related species [11], [12].
Metabolic diversification across plants different taxa (from algae to angiosperms) and their pathway studies may further provide insights for the potential discovery of novel specialized metabolic processes. Genes coding for specialized metabolic functions, their proliferations and physical clustering within the genome when compared with their relative primary metabolic counterparts, some important observations were made [13]. Primary metabolism related reactions, such as carbohydrate metabolism were enriched in early land plant, hormone-related reactions were enriched in algae, whereas angiosperms were enriched for specialized metabolism reactions (amino acid, carbohydrate and nucleotide metabolism reactions significantly underrepresented in angiosperm). In specialized metabolism, gene duplication such as whole-genome duplications (WGDs) and local (tandem) duplication (LDs) play an important role for increase gene content in genomes. For example in Arabidopsis, specialized metabolic genes were significantly enriched in LD genes, but other suggested that whole genome duplications contributed significantly to these metabolic pathways [14]. In soybean and sorghum species, significant depletions of WGD-derived specialized metabolic genes were observed, but displayed significant enrichment in LD-derived specialized metabolic genes [15]. These local duplications of genes appeared to affect a number of flavonoid related genes in each species. Gene clustering also plays a major role in specialized metabolic events, discussed above little bit for the medicinal plant C. roseus. For soybean, sorghum and Arabidopsis, it was shown that approximately one-third of the metabolic genes and one-fifth of the genes in rice were situated in clusters. The Arabidopsis and soybean clusters were significantly enriched for specialized metabolic genes. Clustered genes in Arabidopsis were enriched in phenylpropanoid and terpenoid metabolism, and clustered genes in sorghum were enriched in terpenoid metabolism but the enrichment pattern differed in soybean. The differing results among the species suggest that specialized metabolic gene clusters are a product of independent mechanism rather than a broad mechanism underlying specialized metabolism evolution.
Furthermore, additional research for digging out medicinally important plant metabolic pathways involves various processes including i) gene cloning approaches, ii) RNA interference (RNA) i), and iii) virus-induced gene silencing (VIGS) technology. i) Gene cloning approaches involves protein purification, protein sequencing, and use of protein sequence to clone desired genes. A number of genes involved in the biosynthesis of monoterpenoid indole alkaloids (MIAs) vindoline, camptothecin and catharanthine have been identified and most of the steps in taxol and morphine biosynthesis have been elucidated. Unfortunately, this approach is slow and tedious and most importantly, this requires random testing of many members of the gene family, together with substrate availability, to determine whether a candidate gene is involved in the target pathway [16]. ii) RNA interference is a biological mechanism that eliminates targeted mRNAs through homology dependent gene silencing and has been successfully used in the transformation of plants through the introduction of short double stranded RNA that interferes with the expression of the targeted gene. Transgenic poppies were created through this technology which accumulates the benzylisoquinoline alkaloid reticuline rather than the narcotic morphine [17]. iii) Virus-induced gene silencing technology has been shown to be useful for an increasing number of plant hosts and is particularly useful for plants that are difficult to transform [18]. VIGS technology has identified genes involved in benzylisoquinoline alkaloid (BIA) biosynthesis and has helped identify the two reactions involved in the consecutive O-demethylation of thebaine to codeine and codeine to morphine in opium poppy [19]. Furthermore, although tropane alkaloids (hyoscyamine, atropine and scopolamine) and their biosynthetic pathways have been well characterized, testing candidate genes encoding cytochrome P450s (CYP P450) of Hyoscyamus niger, representing tropane alkaloid pathway-enriched transcripts with VIGS technology identified a CYP80F1 family member that when silenced showed preferential accumulation of littorine rather than hyoscyamine [20]. MIA pathway genes from Catharanthus roseus, though mostly have been characterized, the successful VIGS-mediated suppression of 16-methoxy-2,3-dihydro-3-hydroxytabersonine N-methyltransferase (NMT) in C. roseus and targeted metabolic profiling of NMT-VIGS-silenced plants showed preferential accumulation of NMT substrate 16-methoxy-2,3-dihydro-3-hydroxytabersonine rather than the downstream MIA vindoline [21], [22]. VIGS technology may thus be used as a gene discovery tool, and should greatly speed up the discovery of the remaining MIA pathway genes in C. roseus and other medicinal herbs.
3. Identification of unknown pathways/enzymes for new drug discovery
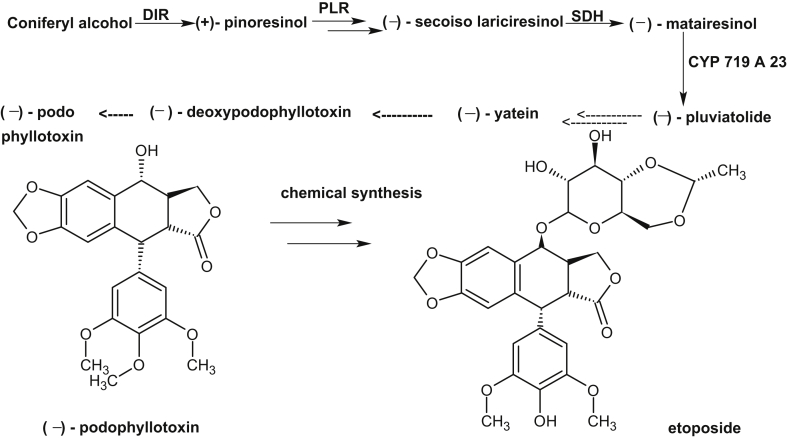
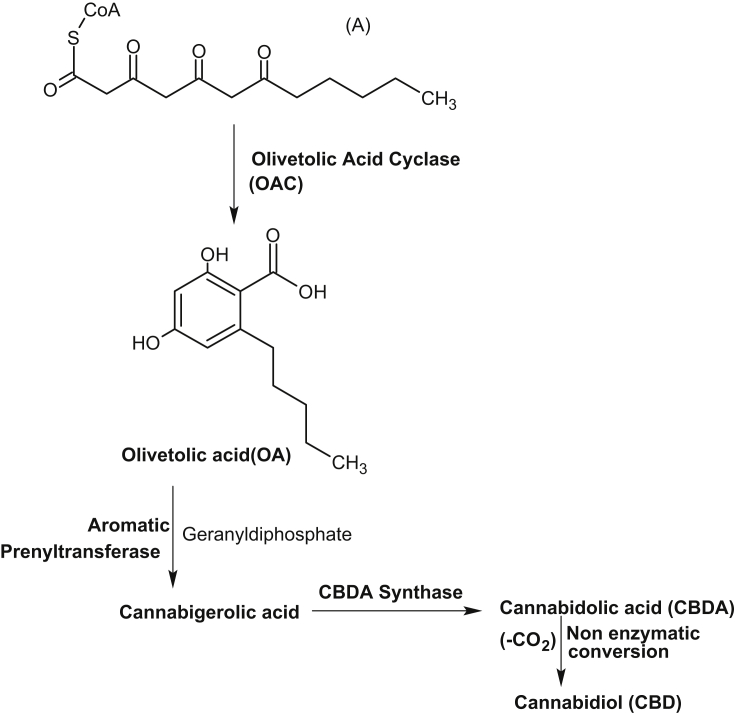
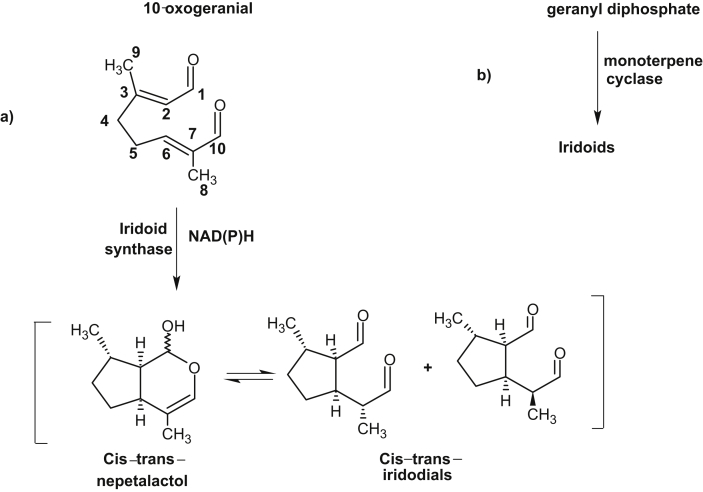
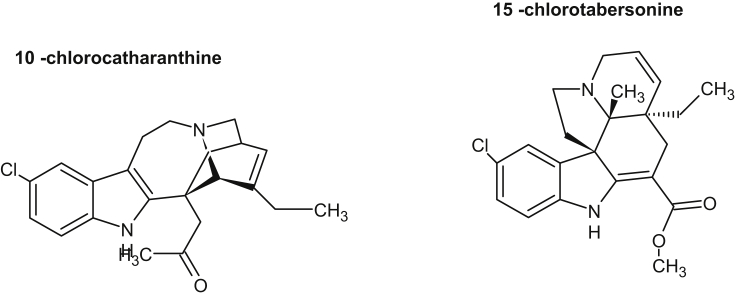
More than one-third of human drugs have originated from products synthesized by plant metabolism. Understanding the origins and vast diversification of plant metabolism has been a long-standing goal in plant biology. Since, only a fraction of the vast chemical biodiversity of the plant world has been explored, its worth stressing on high-throughput sequencing technology to examine increasing number of species for the speedy discovery of previously unknown enzymes and pathways for the production of new medicines. Till date, very little is known about biosynthetic genes of clinically used existing drugs which prevents access to engineered hosts for their production. Very few complete pathways exist, and only three-artemisinic acid and the benzylisoquinoline alkaloids [23], [24], and monoterpenoid indole alkaloids [25], [26] have heterologous host for industrial production. Now, the availability of inexpensive large-scale sequencing tools, together with metabolomic and proteomic information will help to identify candidate genes involved in the biosynthesis of biologically active metabolites. The Medicinal Plant Genomics Consortium and genome-guided investigation [27], [50], the Medicinal Plant Transcriptome Project [28], the 1000 Green Plant Transcriptome Project [29] etc. will add to identify biosynthetic pathways and their evolution in plants. These databases may be specially useful for new pathway discovery if a number of them are found as operon like gene clusters as described in barley, rice [30] and in opium poppy, where a10-gene cluster involved in the biosynthesis of the antitumor alkaloid noscapine was located over 401 kb of genomic sequence [31]. Recently, using transcriptome data from the plant mayapple (Podophyllum hexandrum) and selecting candidate genes to combinatorially express in tobacco (Nicotiana benthamiana), six pathway enzymes of podophyllotoxin pathway to etoposide aglycone were identified [32]. Podophyllotoxin is the natural product precursor of the chemotherapeutic ‘unnatural’ anticancer etoposide, yet only part of its biosynthetic pathway is known (Fig. 1, modified from [32]). By coexpressing altogether 10 genes in tobacco, the pathway to etoposide aglycone, a naturally occurring lignan and intermediate precursor of etoposide was reconstituted. These works not only shows the expression of genes of etoposide precursor in a different plant species but also circumvent the need for cultivation of mayapple. Identification of another plant enzyme, olivetolic acid cyclase, a polyketide cyclase-like enzyme from transcriptome data of glandular trichomes on female cannabis flowers suggests that these cyclases may play an overlooked role in generating plant chemical diversity (Fig. 2, modified form of [34]). Humans have used Cannabis sativa L. (marijuana) as a medicinal and psychoactive herbal drug, and now it is the most widely consumed illicit drug worldwide [33]. Its unique effects are due to the presence of cannabinoids, and the first intermediate in the cannabinoid biosynthetic pathway is proposed to be olivetolic acid (OA), that forms the polyketide nucleus of the cannabinoids. During searching for polyketide cyclase-like enzymes, that could assist in OA cyclization, the enzyme, olivetolic acid cyclase was discovered [34]. Continued search on new enzymatic pathways and compounds having wide range of pharmacological activities, a plant-derived iridoid biosynthetic pathway enzyme, iridoid synthase was discovered [35]. To identify iridoid synthase, recently available transcriptomic data was used from a medicinal plant, Catharanthus roseus that produces a variety of iridoid-derived monoterpene indole alkaloids, including the anticancer alkaloid vinblastine [36]. The iridoids comprise a large family of distinctive bicyclic monoterpenes that possess anticancer, anti-inflammatory and anti-bacterial activities [37], [38], [39]. During iridoid cyclization step, iridoid synthase uses linear monoterpene 10-oxogeranial as substrate which is in contrast to all known monoterpene cyclases that uses geranyl diphosphate as substrate, and the enzyme synthase probably couples an initial NAD(P)H-dependent reduction via a Diels-Alder cycloaddition or a Michael addition (Fig. 3, modified from [36]). This work not only suggests alternative biochemical mechanism for the biosynthesis of cyclic terpenes but also enable the large-scale heterologous production of iridoids in plants. Furthermore, using medicinal plant metabolic pathway enzymes other works are carried out for new drug discovery. For example, through the use of recombinant technology, combining enzyme engineering, genes from microorganisms and plant genetic engineering, search for new classes of compounds and their potentials as new drugs were carried out. For this, an increasing number of enzymes of genes involved in the biosynthesis of plant medicinal products have been crystallized. From the information of the crystal structure of strictosidine synthase, mutants having catalytic activity for broader substrates, together with soil bacteria halogenases were expressed in Catharanthus hairy roots. Transformed hairy roots made chlorinated tryptophan, which was converted via decarboxylase reaction into chlorotryptamine derivatives and into chlorinated monoterpenoid indole alkaloids including 10-chlorocatharanthine and 15-chlorotabersonine [ 40](Fig. 4). This work showed that plant can serve as chemical factories to produce many non-natural monoterpenoid indole alkaloids that can be used to produce new monoterpenoid indole alkaloid-based drugs.
Fig. 1.
Biosynthetic pathway of podophyllotoxin in P. hexandrum. Chemical conversion of anticancer etoposide from podophyllotoxin is also shown. Uncharacterized steps in the pathway are indicated by dashed lines. DIR: dirigent protein, PLR: pinoresinol lariciresinol reductase, SDH: secoisolariciresinol dehydrogenase CYP: cytochrome P 450.
Fig. 2.
Showing Olivetolic Acid Cyclase in the biosynthesis of cannabinoids, a unique biosynthetic route to plant polyketides. The enzyme cyclase catalyzes the formation of OA from hexanoyl CoA plus malonyl CoA (A). OA then converted to cannabigerolic acid and then to major cannabinoids, CBDA through enzymatic conversions. Finally, CBDA upon decarboxylation produces their neutral form CBD, cannabidiol.
Fig. 3.
Iridoid synthase from C. roseus in alternate biochemical mechanism of iridoid biosynthesis. a) The synthase uses monoterpene 10 oxogeranial as substrate and NAD(P)H reduction step for cyclization, whereas b) monoterpene cyclases uses geranyl diphosphate as substrate in the usual process of cyclization.
Fig. 4.
Valuable secondary metabolites produced by pathway engineering in plants. Chlorinated monoterpenoid indole alkaloids are generated from transformed hairy roots of Catharanthus.
Thus, the creation of new pathways in plants and the introduction of new reactions [40] or suppression of existing ones [22], [41], [42] can significantly affect for randomly generating previously unknown molecules. These could be biosynthetic intermediates from an established pathway, or totally new products could be developed from these intermediates [43]. The development of inexpensive genome sequencing together with proteomic and metabolomic information adds to new pathway discovery and may help to identify gene function across plant species. Such studies will definitely help in future to reveal new biologically active secondary metabolites and will take care of natures plant biodiversity for new drug discovery.
4. Concluding remarks
There are now slogans that with tens of thousands of plant species on earth, we are blessed with an enormous wealth of herbal medicine originated from nature and hence under green therapy cover and with no side effect, we can take it in the long term and so forth. Therefore, more and more attention in the field of drug discovery has been focused on the herbal medicine, considering low success rate, high cost and long time of conventional drug development. Plants display an immense diversity of specialized metabolites many of which have been important to humanity as medicines, flavors, fragrances etc. However, only a small fraction of the vast diversity of plant metabolism has been explored. For the speedy discovery of unknown metabolic pathways/enzymes, interdisciplinary research on herbal genomics together with large-scale sequencing tools and metabolomics and proteomics is absolutely necessary. We hope, such studies commits to reveal new biologically active secondary metabolites, making use of vast aspects of plant biodiversity for new drug discovery.
Conflicts of interest
The author declared no conflict of interest with respect to the authorship and/or publication of this article.
Acknowledgments
The author gratefully acknowledges the overwhelming support from American Center Library, Kolkata, and National Library, Kolkata.
References
- 1.Chen S., Song J., Sun C., Xu J., Zhu Y., Verpoorte R., Fan T.-P. Herbal genomics: examining the biology of traditional medicines. Science. 2015;347:527–529. [Google Scholar]
- 2.Patwardhan B., Warude D., Pushpangadan P., Bhatt N. Ayurveda and traditional Chinese medicine: a comparative overview. Evid Based Comple. Alter Med. 2005;2:465–473. doi: 10.1093/ecam/neh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modak M., Dixit P., Londhe J., Ghaskadbi S., Devasagayam T.P.A. Indian herbs and herbal drugs used for the treatment of diabetes. J. Clin. Biochem. Nutr. 2007;40:163–173. doi: 10.3164/jcbn.40.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luca V.D., Salim V., Masada S., Yu F. Mining the biodiversity of plants: a revolution in the making. Science. 2012;336:1658–1661. doi: 10.1126/science.1217410. [DOI] [PubMed] [Google Scholar]
- 5.Chang C., Bowman J.C., Meyerowitz E.M. Field guide to plant model system. Cell. 2016;167:325–339. doi: 10.1016/j.cell.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brower V. Back to Nature: extinction of medicinal plants threatens drug discovery. J. Natl. Cancer Inst. 2008;100:838–839. doi: 10.1093/jnci/djn199. [DOI] [PubMed] [Google Scholar]
- 7.Reymond P., Weber H., Damond M., Farmer E.E. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermsmeier D., Schittko U., Baldwin I.T. Molecular interactions between the specialist herbivore Manduca sexta and its natural host Nicotiana attenuate. I. Large- scale changes in the accumulation of growth- and defense- related plant mRNAs. Plant Physiol. 2001;125:683–700. doi: 10.1104/pp.125.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S., Xu J., Liu C. Genome sequence of the model medicinal mushroom Ganoderma Lucidum. Nat. Commun. 2012;3:913–922. doi: 10.1038/ncomms1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giddings L.A. A stereoselective hydroxylation step of alkaloid biosynthesis by a unique cytochrome P450 in Catharanthus roseus. J. Biol. Chem. 2011;286:16751–16757. doi: 10.1074/jbc.M111.225383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rastogi S., Kalra A., Gupta V. Unravelling the genome of Holy basil an incomparable“elixir of life” of traditional Indian medicine. BMC Genom. 2015;16:413–431. doi: 10.1186/s12864-015-1640-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rastogi S., Meena S., Bhattacharya A. De novo sequencing and comparative analysis of holy and sweet basil transcriptomes. BMC Genom. 2014;15:588–603. doi: 10.1186/1471-2164-15-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chae L., Kim T., Nilo-Poyanco R. Genomic signatures of specialized metabolism in plants. Science. 2014;344:510–513. doi: 10.1126/science.1252076. [DOI] [PubMed] [Google Scholar]
- 14.Kliebemstein D.J. A role for gene duplication and natural variation of gene expression in the Evolution of metabolism. PLoS One. 2008;3:e1838. doi: 10.1371/journal.pone.0001838. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Supplementary materials of Ref. [12] Science. 2014;344:1–31. [Google Scholar]
- 16.Luca V.D., Salim V., Atsumi S.M. Biodiversity of plants: a revolution in the plant biology. Science. 2012;336:1658–1661. doi: 10.1126/science.1217410. [DOI] [PubMed] [Google Scholar]
- 17.Allen R.S., Millgate A.G., Chitty J.A. RNAi-mediated replacement of morphine with the non narcotic alkaloid reticuline in opium poppy. Nat. Biotechnol. 2004;22:1559–1566. doi: 10.1038/nbt1033. [DOI] [PubMed] [Google Scholar]
- 18.Ratcliff F.G., MacFarlane S.A., Baulcombe D.C. Gene silencing without DNA: RNA mediated cross-protection between viruses. Plant Cell. 1999;11:1207–1215. doi: 10.1105/tpc.11.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagel J.M., Facchini P.J. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat. Chem. Biol. 2010;6:273–275. doi: 10.1038/nchembio.317. [DOI] [PubMed] [Google Scholar]
- 20.Li R., Reed D.W., Liu E. Functional genomic analysis of alkaloid biosynthesis in Hyoscyamus Niger reveals a cytochrome P450 involved in littorine rearrangement. Chem. Biol. 2006;13:513–520. doi: 10.1016/j.chembiol.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Roepke J., Salim V., Wu M. Vinca drug components accumulate exclusively in leaf exu-dates of Madagascar periwinkle. Proc. Natl. Acad. Sci. USA. 2010;107:15287–15292. doi: 10.1073/pnas.0911451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liscombe D.K., O'Connor S.E. A virus-induced gene silencing approach to understanding alkaloid metabolism in Catharanthus roseus. Phytochemistry. 2011;72:1969–1977. doi: 10.1016/j.phytochem.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paddon C.J., Westfall P.J., Pitera D.J. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 24.Thodey K., Galanie S., Smolke C.D. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat. Chem. Biol. 2014;10:837–844. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown S., Clastre M., Courdavault V. De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl. Acad. Sci. USA. 2015;112:3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu Y., Easson M.L.A.E., Froese J. Completion of the seven step pathway from taberso- nine to the anticancer drug precursor vindoline and its assembly in yeast. Proc. Natl. Acad. Sci. USA. 2015;112:6224–6229. doi: 10.1073/pnas.1501821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellner A. 2015. Genome sequence of Catharanthus roseus.http://medicinalplantgenomics.msu.edu/ [Google Scholar]
- 28.Transcriptome characterization, sequencing, and assembly of medicinal plants relevant to human health, http://uic.edu/pharmacy/med pl transcriptome/index.html.
- 29.Plants 1000 (one KP or 1 KP) gene sequencing data, www.onekp.com.
- 30.Chu H.Y., Wegel E., Osbourn A. From hormones to secondary metabolism : the emergence of metabolic gene clusters in plants. Plant J. 2011;66:66–79. doi: 10.1111/j.1365-313X.2011.04503.x. [DOI] [PubMed] [Google Scholar]
- 31.Wizer T., Gazda V., He Z. A Papaver somniferum 10-Gene cluster for synthesis of the anticancer alkaloid noscapine. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. [DOI] [PubMed] [Google Scholar]
- 32.Lau W., Sattely E.S. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science. 2015;349:1224–1228. doi: 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.United Nations office on drugs and crime . United Nations office on drugs and crime; Vienna, Austria: 2010. World Drug Report 2010. [Google Scholar]
- 34.Gagne S.J., Stout J.M., Liu E. Identification of olivetolic acid cyclase from Cannabissativa reveals a unique catalytic route to plant polyketides. Proc. Natl. Acad. Sci. USA. 2012;109:12811–12816. doi: 10.1073/pnas.1200330109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geu-Flores F., Sherden N.H., Courdavault V. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature. 2012;492:138–142. doi: 10.1038/nature11692. [DOI] [PubMed] [Google Scholar]
- 36.Dinda B., Chowdhury R., Mohanta D. Naturally occurring iridoids, secoiridoids and their bioactivity. Chem. Pharm. Bull. (Tokyo) 2009;57:765–796. doi: 10.1248/cpb.57.765. [DOI] [PubMed] [Google Scholar]
- 37.Dinda B., Debnath S., Banik R. Naturally occurring iridoids and secoiridoids. Chem. Pharm. Bull. (Tokyo) 2011;59:803–833. doi: 10.1248/cpb.59.803. [DOI] [PubMed] [Google Scholar]
- 38.Dinda B., Debnath S., Harigaya Y. Naturally occurring secoiridoids and bioactivity natu- rally occurring iridoids and secoiridoid. Chem. Pharm. Bull. (Tokyo) 2007;55:689–728. doi: 10.1248/cpb.55.689. [DOI] [PubMed] [Google Scholar]
- 39.Tundis R., Loizzo M.R., Menichini F. Biological and pharmacological activities of iridoids: recent developments. Med. Chem. 2008;8:399–420. doi: 10.2174/138955708783955926. [DOI] [PubMed] [Google Scholar]
- 40.Runguphan W., Qu X., O'Connor S.E. Integrating carbon-halogen bond formation into medicinal plant metabolism. Nature. 2010;468:461–464. doi: 10.1038/nature09524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hileman L.C., Drea S., Martino G. Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy) Plant J. 2005;44:334–341. doi: 10.1111/j.1365-313X.2005.02520.x. [DOI] [PubMed] [Google Scholar]
- 42.Wege S., Scholz A., Gleissberg S. Highly efficient virus-induced gene silencing in California poppy (Eschscholzia californica): an evaluation of VIGS as a strategy to obtain functional data from non-model plants. Ann. Bot. (Lond) 2007;100:641–649. doi: 10.1093/aob/mcm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Facchini P.J., Bohlmann J., Covello P.S. Synthetic biosystems for the production of high-value plant metabolites. Trends Biotech. 2012;30:127–131. doi: 10.1016/j.tibtech.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Chen S., Sun Y.Z., Xu J. Strategies of the study on herb genome program. Yao Xue Xue Bao. 2010;45:807–812. [PubMed] [Google Scholar]
- 45.Chen S., Xu L.X., Guo Q.L. An introduction to the medicinal plant genome project. Front. Med. 2011;5:178–184. doi: 10.1007/s11684-011-0131-0. [DOI] [PubMed] [Google Scholar]
- 46.Mahajan R.T.C.M. Phyto-pharmacology of Ziziphus Jujuba Mill-a plant review. Pharmacol. Rev. 2009;3:320–329. [Google Scholar]
- 47.Shivaraj Y., Govind S., Jogaiah S. Functional analysis of medicinal plants using system biology approaches. Int. J. Pharm. Pharmaceut. Sci. 2015;7:41–43. [Google Scholar]
- 48.He Y., Xiao H., Deng C. The complete chloroplast genome sequences of the medicinal plant Pogostemon cablin. Int. J. Mol. Sci. 2016;17:820–830. doi: 10.3390/ijms17060820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qian J., Song J., Gao H. The complete chloroplast genome sequences of the medicinal plant Salvia miltiorrhiza. PLoS One. 2013;8:e57607. doi: 10.1371/journal.pone.0057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kellner F., Kim J., Clavijo B.J. Genome-guided investigation of plant natural product biosynthesis. Plant J. 2015;82:680–692. doi: 10.1111/tpj.12827. [DOI] [PubMed] [Google Scholar]
- 51.Li Y., Xu C., Lin X. De novo assembly and characterization of the fruit transcriptome of Chinese Jujuba(Zizipus Jujuba Mill) using 454 pyrosequencing and the development of novel trinucleotide SSR markers. PLoS One. 2014 doi: 10.1371/journal.pone.0106438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu H., Song J., Luo H. Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol. Plant. 2016;9:949–952. doi: 10.1016/j.molp.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]