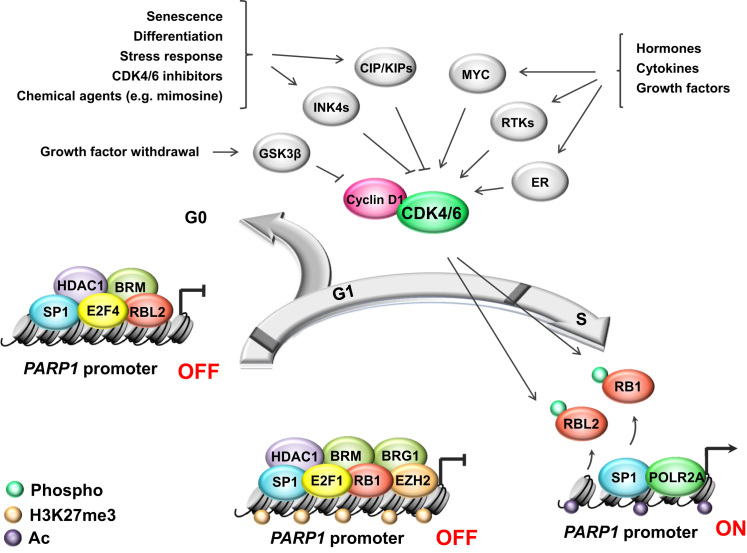
Fig. 1.
Cell cycle progression dictatesPARP1transcription via growth factors/inhibitors-G1/G0-CDK4/6-RBs axis. Cell cycle machinery is controlled by external signals in order to adapt cells to environmental requirements and conditions. Stimulation of receptor tyrosine kinases (RTKs), MYC protooncogene or estrogen receptor (ER) in response to peptide and non-peptide growth-promoting agents activates cyclin-dependent kinase 4 and 6 (CDK4/6), which associate with cyclin D1 and phosphorylate retinoblastoma proteins (RB1, RBL2). This modification keeps retinoblastoma proteins released from promoters of PARP1 and cell cycle promoting genes, thereby allowing active gene transcription and enabling cell transition from G1 to S phase. Upon cell growth arrest in G1 or cell cycle exit to G0, CDK4/6 inhibition results in hypophosphorylation of retinoblastoma proteins, their binding to E2F-driven gene promoters and recruitment of chromatin remodelers, which are capable of inactivating gene expression by removing transcription-promoting indicators and/or inserting transcription-inhibiting histone modification(s). It leads to an increase in nucleosome density and chromatin condensation. Notably, composition of the repressive complex varies between cells arrested in G1 and in G0. Limiting PARP1 expression in G0 is achieved solely by histone deacetylase 1 (HDAC1) for histone deacetylation, while in G1 HDAC1 additionally requires PRC2 (polycomb repressor complex 2) activity and trimethylation of H3K27 by enhancer of zeste homolog 2 (EZH2) to repress PARP1 transcription. Cell cycle arrest in G2 does not affect the mRNA and protein levels of PARP1.