Fig. 4.
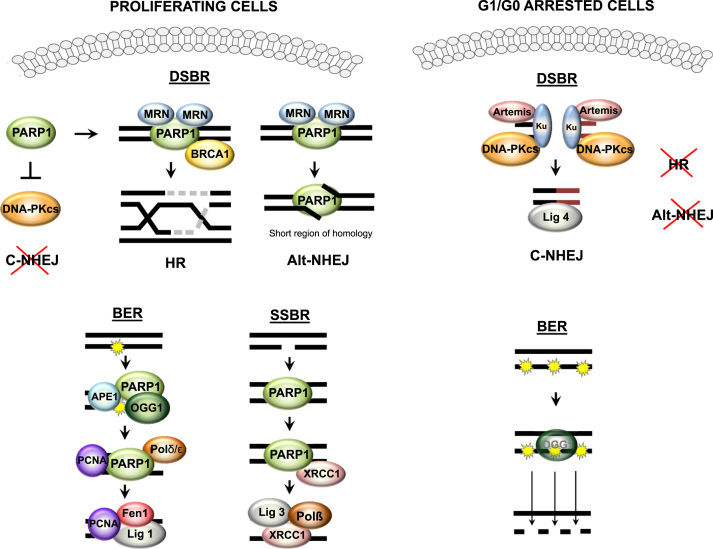
PARP1 regulates repair of oxidative DNA damages. The cell decision on involvement of particular repair system depends on the type of DNA damage and is strongly related to cell cycle progression. For double strand breaks, the cell is equipped with three repair systems: homologous recombination (HR), classical non-homologous end joining (C-NHEJ) or alternative non-homologous end joining (Alt-NHEJ). Proliferating cells, with high PARP1 levels, make use mainly of two mechanisms: HR and/or Alt-NHEJ, which assure accurate and error-free repair of double strand breaks since they use a replicated DNA template to reconstruct the missing fragment with high fidelity. These two pathways rely on the recognition of detrimental lesions by PARP1, which recruits other proteins to the affected sites: first MRN complex (consisting of MRE11, Rad50, Nbs1), an initiator of repair, then MRE11 determines the composition of proteins for each repair mechanism (HR or NHEJ). The low C-NHEJ involvement in repair is achieved by high expression of PARP1, an inhibitor of DNA-dependent protein kinases (DNA-PKcs), which is crucial for C-NHEJ progression. In G1/G0 arrested cells (deficient in HR and Alt-NHEJ), low expression of PARP1 allows recruitment of the Ku70/80 heterodimer to double strand breaks and sites of DNA-PKcs activation. Repair machinery such as nuclease Artemis, DNA ligase IV and XRCC4 further process the damaged DNA and directly ligate DNA ends, leading to the irreversible loss of genetic material. Base excision repair (BER) is a substantial pathway to repair oxidized bases. The damaged base is recognized and removed by OGG1 glycosylase, which requires PARP1 for proper and efficient functioning. AP endonuclease (APE1) is binds to apurynic sites and produces single-strand breaks (SSBs), which again involve PARP1 in the BER machinery at a later repair step. DNA nick-induced poly-ADP-ribosylation facilitates PARP1 interaction with DNAPδ/ε and PCNA, which further govern repair machinery. Single strand breaks resulting from direct oxidant action also need PARP1 activity to be repaired. Poly-ADP-ribose polymers recruit XRCC1, then the remaining SSBR machinery comprising DNA polymerase (DNAP) and DNA ligase III, which also can be PARylated by PARP1. PARP1 deficiency in growth-arrested cells substantially impairs both pathways. Of note, inhibition of OGG1 in combination with G1-blockade leads to further accumulation of single strand breaks.