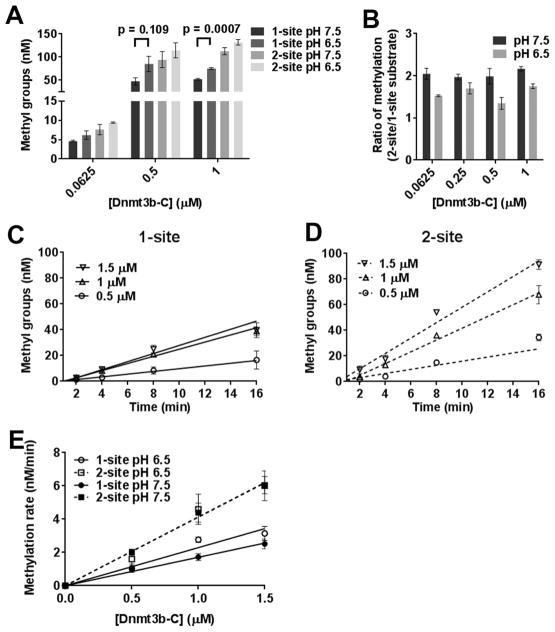
Figure 6.
pH sensitivity of Dnmt3b-C processive catalysis. (A) Methylation at one- and two-site substrates by Dnmt3b-C was measured for 1 h at pH 6.5 or 7.5. (B) Ratio of the methylation levels calculated as the fold change at two-site vs one-site substrate. (C and D) Time course of DNA methylation by Dnmt3b-C at pH 6.5 for one- and two-site substrates. For each substrate, the time course of DNA methylation reaction was determined in the presence of 0.5–2 μM Dnmt3b-C. The reaction was started by the addition of enzyme, and the data were fitted by linear regression, which was weighted by 1/Y2. (E) The calculated methylation rates at pH 6.5 were plotted vs enzyme concentration and compared to those at pH 7.5. Data are the average ± the standard error of the mean (n ≥ 4 independent experiments).