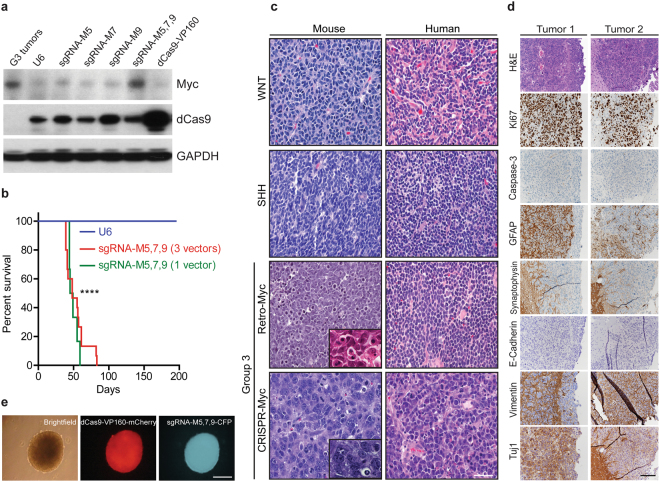
Figure 2.
Characterization of Myc activation in CRISPR-Myc MBs. (a) Detection of Myc and dCas9 protein by immunoblotting of Trp53-null neurosphere cells stably expressing pT2K-CAGGS-dCas9-VP160-T2A-Puro-mCherry and sgRNA-M5, M7, M9 as single guides and as a pool sgRNA-M5,7,9. G3 tumors were used as positive control for Myc levels. GAPDH was used as loading control. Full blots are presented in Supplementary Fig. S5. (b) Kaplan-Meier survival curves for mice orthotopically transplanted with engineered Trp53-null neurospheres stably expressing pT2K-CAGGS-dCas9-VP160-T2A-Puro-mCherry and the pool expressing sgRNA-M5, M7 and M9 (n = 15) or a single vector construct expressing all three guides sgRNA-M5,7,9 (n = 6). Median survival time for mice bearing tumors: 48 days post-transplantation. Negative control cells expressed pT2K-CAGGS-dCas9-VP160-T2A-Puro-mCherry without sgRNAs (n = 5). ****p < 0.0001. (c) Hematoxylin and Eosin (H&E) staining of mouse and human WNT, SHH, G3 (Retro-Myc), and G3 (CRISPR-Myc) MBs showing large cell anaplastic features typify G3 MBs as shown in inserts. Scale bar, 100 µm. (d) Sections of CRISPR-Myc tumors immunostained with antibodies to H&E, Ki67, Caspase-3, GFAP, Synaptophysin, E-Cadherin, Vimentin, and Tuj1. Scale bar, 200 µm. (e) Tumors harvested from (b) cultured as neurospheres showed stable expression of dCas9-VP160 (mCherry) and sgRNA-M5, 7, 9 (CFP). Scale bar, 100 µm.