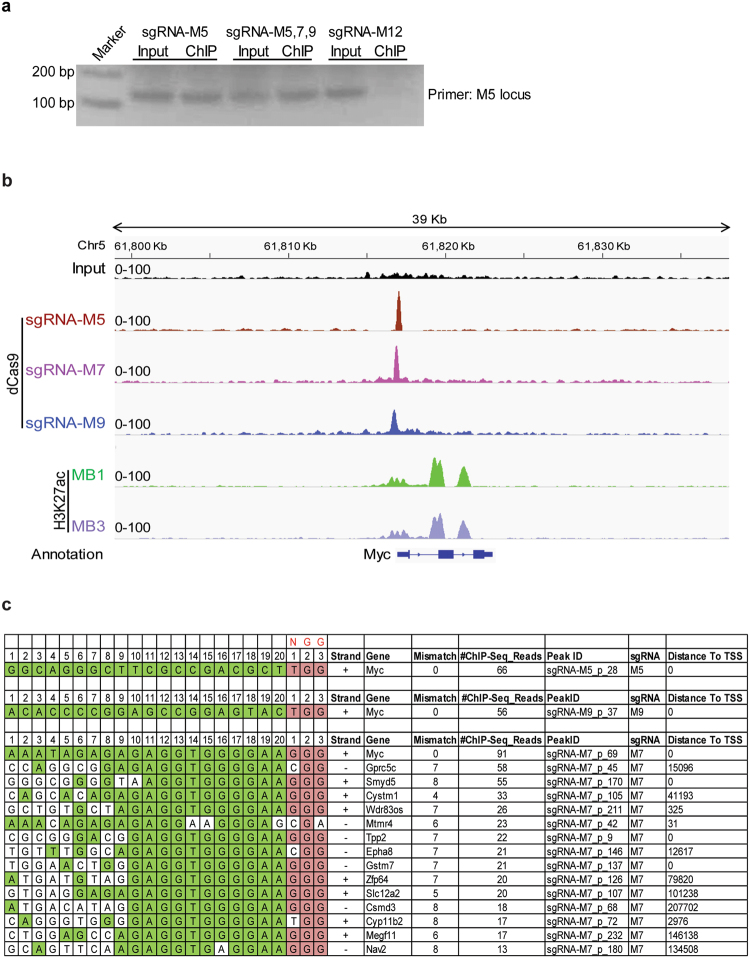
Figure 4.
Validation of sgRNAs specific binding to the endogenous Myc promoter in CRISPR activation system. (a) Primers flanking the sgRNA-M5 binding motif was used for ChIP-PCR in Trp53-null neurosphere cells infected with Lenti-dCas9-VP64-T2A-GFP and Lenti-U6-sgRNA-RFP with sgRNA-M5 or combination with sgRNA-M5, 7, and 9. sgRNA-M12 was used as a negative control. Full agarose gel is presented in Supplementary Fig. S6. (b) Enrichment of sgRNA-M5, M7 and M9 binding to pre-designated loci in Trp53-null neurosphere cells revealed by ChIP-Seq using dCas9 antibody. ChIP-Seq of H3K27ac on mouse G3 MB cell lines MB1 and MB3 illustrated to define open chromatin status of the endogenous Myc locus. (c) Mapping reads from ChIP-Seq of dCas9 used to characterize on-target (M5, M9) and off-target (M7) sequences.