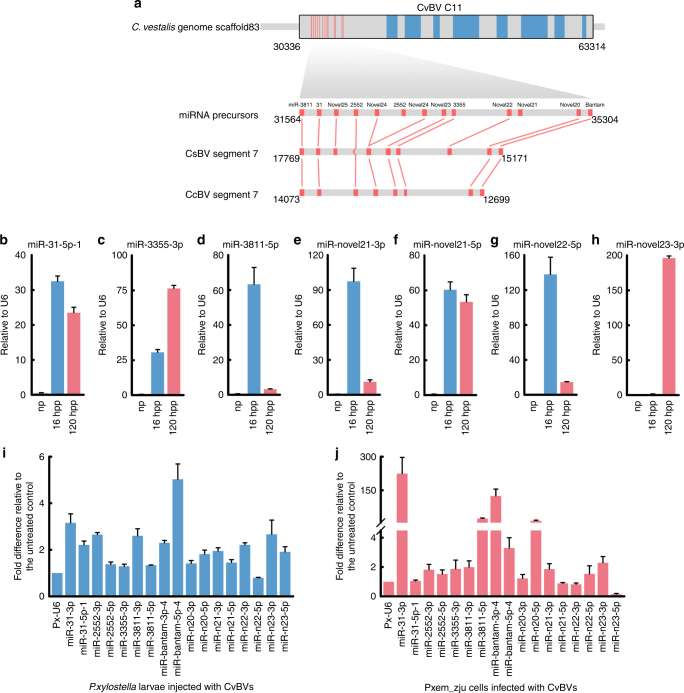
Fig. 2.
CvBV-derived miRNAs and their expression in P. xylostella. a miRNAs precursors located in CvBV segment C11. The upper bar schematically shows segment C11 in C. vestalis genome scaffold 83. Red vertical lines represent miRNA precursors while blue vertical bars identify predicted protein coding sequences. Below the upper bar is shown the domain of segment C11 that encodes miRNA precursors together with schematics showing miRNA precursors identified in CsBV segment 7 and CcBV segment 7 (Results). b–h RT-qPCR analysis of eight CvBV-derived miRNAs in P. xylostella normalized to P. xylostella U6 snRNA. np: non-parasitized P. xylostella, hpp: hours post parasitized. Ct values for undetected samples are considered as 40. i RT-qPCR analysis of 17 CvBV-encoded miRNAs in P. xylostella larvae injected 24 h earlier with 0.1 FEs of CvBV particles. P. xylostella were injected during third instars. Relative expression of CvBV-encoded miRNAs was normalized to the P. xylostella U6 snRNA. j RT-qPCR analysis of 17 CvBV-encoded miRNAs in Pxem-ZJU cells (5 × 106 cells) 24 h post infection with 20 FEs of CvBV particles. The expression of CvBV-encoded miRNAs was normalized to P. xylostella U6 snRNA. Results shown are mean relative abundance ± s.e.m. of each miRNA from three independent biological replicates