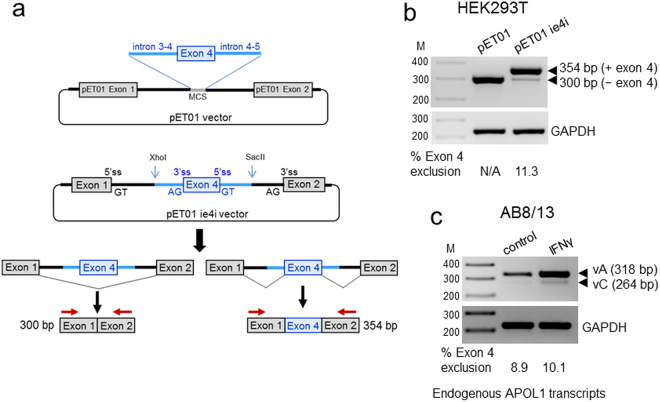
Figure 2.
Generation of a splicing reporter pETie4i minigene for the analysis of the APOL1 exon 4 splicing. (a) APOL1 exon 4 and the flanking intron 3–4 sequence (includes 197 bp of full 2,083 bp) and a full intron 4–5 (182 bp), were PCR-amplified from genomic DNA of AB8/13 podocytes and cloned into the XhoI/SacII sites of the pET01 vector to create the pETie4i minigene construct. Two alternatively spliced transcripts can be produced by the pETie4i, transcript lacking exon 4 (300 bp) and with exon 4 (354 bp). MCS, multicloning site; ss, splice site. (b) pET01 and pETie4i constructs were transfected into HEK293T and 24 h later, total RNA was reverse transcribed to generate cDNA, which was subsequently amplified using a set of primers specific for pET01 exon 1 and exon 2 (red arrows) or primers specific for GAPDH (control). N/A, not applicable. (c) Endogenous APOL1 transcripts expressed in IFNγ-stimulated AB8/13 podocytes were analyzed by RT-PCR using a set of primers p1_3/p6 (see Fig. 1c) designed to amplify fragments of APOL1 vA and vC. The PCR products were separated on 2% agarose gel and stained with ethidium bromide. Predicted PCR product sizes (bp) are shown. Percentage of transcripts with or without exon 4 was calculated from densitometric scanning of the bands. Transcripts generated from transfected pETie4i (b) and endogenous APOL1 transcripts expressed in AB8/13 podocytes (c) show a similar pattern of exon 4 exclusion (11.3 vs.10.1%), indicating that pETie4i correctly reproduces the splicing pattern of endogenous APOL1 transcripts. M, DNA markers (bp). The gel images (b and c) were cropped. The full gel images are shown in Supplementary Fig. 2.