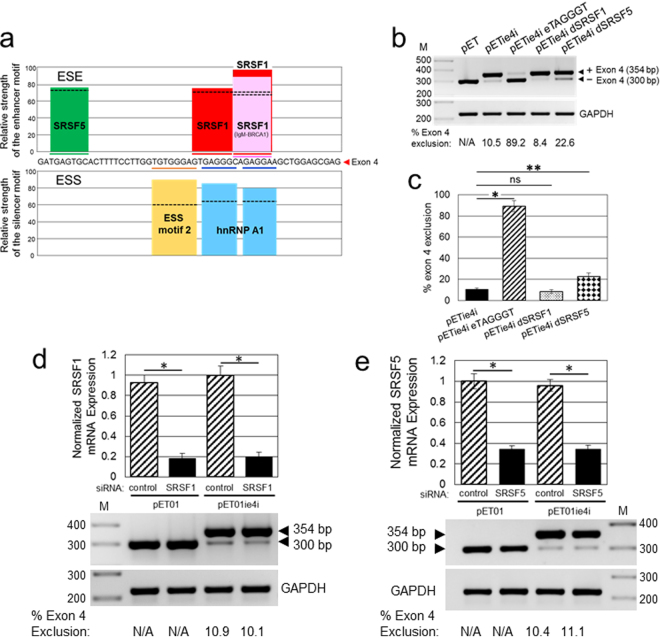
Figure 3.
Effect of SRSF1 and SRSF5 on exon 4 skipping. (a) Identification of putative exonic splicing enhancer (ESE) and exonic splicing silencer (ESS) motifs in the APOL1 exon 4. The upper panel presents ESE motifs identified by ESEfinder 3.0. Scores above the threshold (black dotted lines) were found for SRSF1 and SRSF5 proteins. The lower panel shows ESS motifs predicted by Human Splicing Finder 3.0 (HSF3.0). Two major ESS motifs (ESS motif 2 and hnRNP A1) were identified. (b) Mutation in SRSF1 binding motif (GGCAG > ATATA, pETie4i dSRSF1), predicted by ESEfinder 3.0 to abolish SRSF1 binding motifs in exon 4, does not affect exon 4 splicing. Mutation in SRSF5 binding motif in exon 4 (TGAGTGC > TATATGC, pETie4i dSRSF5), which abolishes predicted by ESEfinder 3.0 binding motif for SRSF5, enhances moderately exon 4 skipping. Minigene with hnRNPA1 consensus motif in exon 4 (pETie4i eTAGGGT) served as positive control for exon 4 skipping. (c) Exon 4 exclusion was calculated from the intensities of RT-PCR bands shown in (b) and was expressed as a percentage of transcripts without exon 4 in a total pool of transcripts with and without exon 4 (100%). HEK293T cells were transfected with indicated minigene constructs and after 24 h, RNA was isolated and subjected to RT-PCR. Values are means ± SD of 3 independent experiments. *P < 0.001, **P < 0.005; ns, not statistically significant. pET, empty minigene control; pETie4i, minigene with exon 4 and adjacent introns. Effect of knockdown of SRSF1 (d) and SRSF5 (e) on exon 4 skipping. HEK293T cells were transfected with SRSF1 siRNA (d), SRSF5 siRNA (e) or control siRNA. After 24 h, the cells were transfected with pET01 or pETie4i minigenes and 24 h later total RNA was collected and subjected to RT-PCR. Values are means ± SD of 3 independent experiments. *P < 0.001. The gel images (b–e) were cropped. The full gel images are shown in Supplementary Fig. 3.