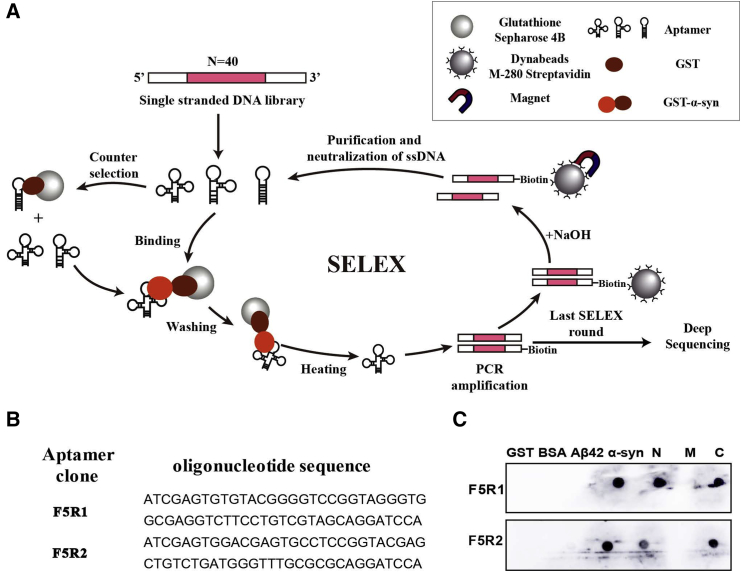
Figure 1.
α-syn Aptamers Were Selected through SELEX
(A) Schematic illustration of the method used for α-syn aptamer selection. GST-tagged α-syn was immobilized on glutathione-sepharose beads. The ssDNA library was incubated with the target beads for binding. Unbound oligonucleotides were washed away, and the bound ones were released by heating at 95°C. The selected binders were amplified by PCR with biotinylated primers. ssDNAs were subsequently purified from the PCR product using streptavidin-coated magnetic beads, resulting in an enriched DNA pool, which was used in the next SELEX round. After the last round, the selected ssDNAs were sequenced by deep sequencing. (B) The aptamer candidates. After deep sequencing, the two sequences with most frequently appearing were selected as the aptamer candidates. (C) Aptamer binding specificity assay by dot blotting. Five microgram samples (α-syn, GST, Aβ42, BSA, and three domains of α-syn) were respectively immobilized onto the nitrocellulose membrane for binding of each aptamer.