Abstract
MicroRNAs are known to be involved in carcinogenesis. Recently, microRNA-372 (miR372) has been proven to play a substantial role in several human cancers, but its functions in liver cancer remain unclear. Herein, our results demonstrate that miR372 accelerates growth of liver cancer cells in vitro and in vivo. Mechanistically, miR372 enhances expression of Y-box-binding protein 1 (YB-1) by targeting for phosphatase and tensin homolog (PTEN) directly and consequently promotes phosphorylation of YB-1 via HULC looping dependent on ERK1/2 and PTEN. In particular, HULC knockdown or PTEN overexpression abrogated this miR372 action. Moreover, miR372 inhibits the degradation of β-catenin dependent on phosphorylation of YB-1 and then enhances the expression and activity of pyruvate kinase M2 isoform (PKM2) by β-catenin-LEF/TCF4 pathway. Furthermore, the loading of LEF/TCF4 on PKM2 promoter region was significantly increased in miR372 overexpressing Hep3B, and thus, glycolytic proton efflux rate (glycoPER) was significantly increased in rLV-miR372 group compared to the rLV group. Moreover, β-catenin knockdown abrogates this function of miR372. Ultimately, miR372 promotes the expression of erbB-2 through PKM2-pH3T11-acetylation on histone H3 lysine 9 (H3K9Ac) pathway. Of significance, both YB-1 knockdown and erbB-2 knockdown abrogate oncogenic action of miR372. Our observations suggest that miR372 promotes liver cancer cell cycle progress by activating cyclin-dependent kinase 2 (CDK2)-cyclin E-P21/Cip1 complex through miR372-YB-1-β-catenin-LEF/TCF4-PKM2-erbB-2 axis. This study elucidates a novel mechanism for miR372 in liver cancer cells and suggests that miR372 can be used as a novel therapeutic target of liver cancer.
Keywords: liver cancer, miR372, YB-1, β-catenin, PKM2, erbB-2, CDK2
Introduction
Recently, microRNA-372 (miR372) has been proven to play a substantial role in several human cancers.1 For example, excessive miR372 promotes metastasis of oral and liver cancer.2 Importantly, miR372 activates the nuclear factor κB (NF-κB) signaling and enhances autophagy in cancer cells.3, 4 Moreover, a miR372/let-7 axis regulates human somatic cell and cancer cell fates.5, 6 Studies indicate that miR372 inhibits the Ras homolog gene family member C (RhoC)7 and p62 in human cancer cells.8 In particular, repressing miR-372 by arsenic sulfide (As4 S4) could inhibit prostate cancer cell proliferation and migration.9 However, the tumor-suppressing roles of miR372 were also found in some cancer cells, possibly via the downregulation of cyclin-dependent kinase 2 (CDK2) and CCNA1.10, 11 Downregulation of ULK1 by miR372 inhibits the survival of human pancreatic adenocarcinoma cells.12 In addition, interleukin-1β (IL-1β) upregulates miR-372 to inhibit spinal cord injury recovery.13 miR-372 acts as a tumor suppressor gene by regulating p65 in prostate cancer (PCa), which may provide a strategy for blocking PCa metastasis.14 Furthermore, circulating miR372 is a novel tumor biomarker for colorectal cancer (CRC) detection.15 Moreover, miR372 suppresses tumor proliferation and invasion by targeting IGF2BP1 in renal cell carcinoma.16 Furthermore, miR372 suppressed the expression of ATAD2, which was highly expressed in hepatocellular carcinoma (HCC) and exerted a proto-oncogene effect in hepatic carcinogenesis.17
In this study, we indicate that miR372 accelerates malignant progression of liver cancer cells. Moreover, miR372 increases CTCF by targeting for phosphatase and tensin homolog (PTEN) and enhances erbB-2 through YB1-β-catenin-pyruvate kinase M2 isoform (PKM2) pathway. This study elucidates a novel mechanism for miR372 in liver cancer cells.
Results
miR372 Accelerates Growth of Liver Cancer Cells
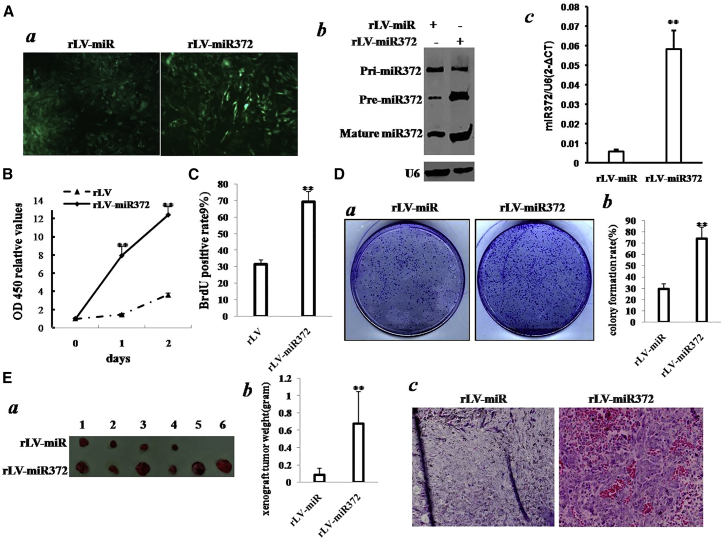
To validate whether miR372 influences malignant growth of human liver cancer cells, we first constructed two stable Hep3B cell lines by infecting with rLV or rLV-miR372. As shown in Figure 1Aa, the green was expressed in two groups. Compared to rLV group, both pre-miR372 and mature miR372 were significantly overexpressed in rLV-miR372 group (Figures 1Ab and 1Ac). As shown in Figure 1B, excessive miR372 significantly increases the growth ability of liver cancer cell Hep3B compared to the control group (p < 0.01). Moreover, the bromodeoxyuridine (BrdU)-positive rate is significantly increased in rLV-miR372 compared to the rLV group (69.01% ± 6.38% versus 31.44% ± 2.53%; p = 0.008415 < 0.01; Figure 1C). Furthermore, we performed colony formation assay and observed a significant increase in colony formation efficiency rate in rLV-miR372 compared to rLV control group (73.93% ± 10.08% versus 29.55% ± 4.38%; p = 0.0089 < 0.01; Figure 1D). Furthermore, our finding also showed rLV did not influence proliferation ability of Hep3B (p > 0.05; Figure S1A) and colony formation ability (33.26% ± 8.43% versus 34.38% ± 3.71%; p = 0.37746 > 0.05; Figure S1A).
Figure 1.
miR372 Accelerates Liver Cancer Cell Growth In Vitro and In Vivo
(A) (a) The photography of the Hep3B cell lines infected with rLV or rLVmiR372. (b) Northern blotting analysis of miR372 in Hep3B cell lines infected with rLV and rLV-miR372 is shown. U6 as internal control is shown. (c) The real-time PCR detection of mature miR372 in Hep3B cell lines infected with rLV and rLV-miR372, respectively, is shown. Each value was presented as mean ± SEM. **p < 0.01. (B) Cell proliferation assay was performed in 96-well format using the CCK8 cells proliferation kit to determine the cell viability as described by the manufacturer. Each sample was assayed in triplicates for 3 days consecutively. Cell growth curve was based on the corresponding relative values of OD450, and each point represents the mean of three independent samples. Data are means of value from three independent experiments; mean ± SEM. **p < 0.01 ; *p < 0.05. (C) Cell BrdU assay is shown. Data are means of value from three independent experiments; mean ± SEM. **p < 0.01; *p < 0.05. (D) (a) The photography of colonies from the cell lines indicated in left is shown. (b) Cell plate colony formation ability assay is shown. Data are means of value from three independent experiments; mean ± SEM. **p < 0.01; *p < 0.05. (E) (a) The photography of the xenograft tumors from Balb/C-null mouse injected with Hep3B cells transfected with rLV and rLV-miR372 subcutaneously at armpit is shown. (b) The xenograft tumor weight (gram) in the two groups is shown. Data were means of value from six BALB/c mice; mean ± SEM; n = 6; *p < 0.05; **p < 0.01. (c) A portion of each xenograft tumor was fixed in 4% formaldehyde and embedded in paraffin, and the micrometers of sections (4 μm) were made for H&E staining (original magnification ×100).
To further explore the effect of miR372 on liver cancer cells in vivo, the two stable Hep3Bs were injected subcutaneously into athymic Balb/C mice. As shown in Figures 1Ea and 1Eb, when miR372 was overexpressed, the xenograft tumor weight increases approximately eight-fold compared to the corresponding control group (0.08333 ± 0.07715 g versus 0.675 ± 0.037284 g; p = 0.00605 < 0.01). Moreover, compared to rLV group, xenograft tumors contain more poorly differentiated cells in rLV- miR372 group (Figure 1Ec). Moreover, we performed the growth assay in vitro and in vivo in liver cancer cell line Huh7. We obtained similar results as Hep3B (Figures S2–S4).
Taken together, these findings demonstrate that miR372 accelerates malignant growth of liver cancer cells.
miR372 Increases the CTCF Expression by Inhibiting PTEN
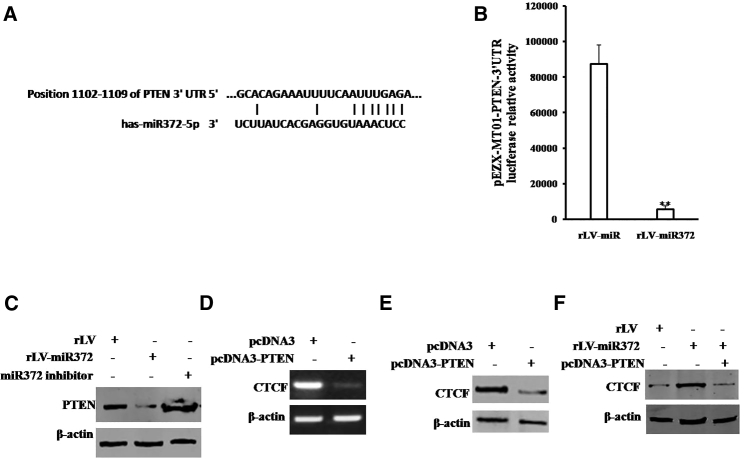
To address whether miR372 could regulate CTCF dependent on PTEN in human liver cancer cells, we first perform the informatics analysis using MirTarget scanning soft and BLAST analysis. As shown in Figure 2A, mature miR372 matches 3′ UTR on PTEN mRNA via eleven-seed sequence. As shown in Figure 2B, the PTEN 3′ UTR luciferase activity was significantly reduced in miR372-overexpressing Hep3B cells compared to control group (5,681 ± 1,999.1 versus 87,488.7 ± 10,652.2; p = 0.00357 < 0.01). Moreover, the expression of PTEN was significantly decreased in miR372-overexpressing Hep3B cells compared to control group. However, the expression of PTEN was significantly increased in miR372-inhibiting Hep3B cells compared to control group (Figure 2C). Furthermore, the transcription of CTCF was significantly decreased in PTEN-overexpressing Hep3B cells compared to control group (Figure 2D). Also, the expression of CTCF was significantly decreased in PTEN-overexpressing Hep3B cells. Strikingly, the expression of CTCF was significantly increased in miR372-overexpressing Hep3B cells compared to control group. However, the expression of CTCF was not significantly altered in miR372-overexpressing plus PTEN-overexpressing Hep3B cells compared to control group (Figure 2F). Collectively, our findings suggest miR372 targets for PTEN 3′ UTR directly and increases CTCF expression by inhibiting PTEN in liver cancer cells.
Figure 2.
miR372 Promotes CTCF by Targeting PTEN Directly in Liver Cancer Cells
(A) Bioinformatics analysis: miR675 targets for human PTENα 3′ UTR. (B) pEZX-MT01-PTEN-3′-UTR luciferase activity assay is shown. Data are means of value from three independent experiments, mean ± SEM. **p < 0.01. (C) Western blotting analysis using anti-PTEN in Hep3B cells infected with rLV and rLV-miR372, respectively, is shown. β-actin as internal control is shown. (D) RT-PCR analysis using CTCF primers in Hep3B cells transfected with pcDNA3 and pcDNA3-PTEN, respectively, is shown. β-actin as internal control is shown. (E) Western blotting analysis using anti-CTCF in Hep3B cells transfected with pcDNA3 and pcDNA3-PTEN, respectively, is shown. β-actin as internal control is shown. (F) Western blotting analysis using anti-CTCF in Hep3B cell lines, including rLV, rLV-miR372, and rLV-miR372 plus transfected pcDNA3-PTEN, is shown.
miR372 Enhances Expression of YB-1 through the Specific DNA Loop Mediated by CTCF in Liver Cancer Cells
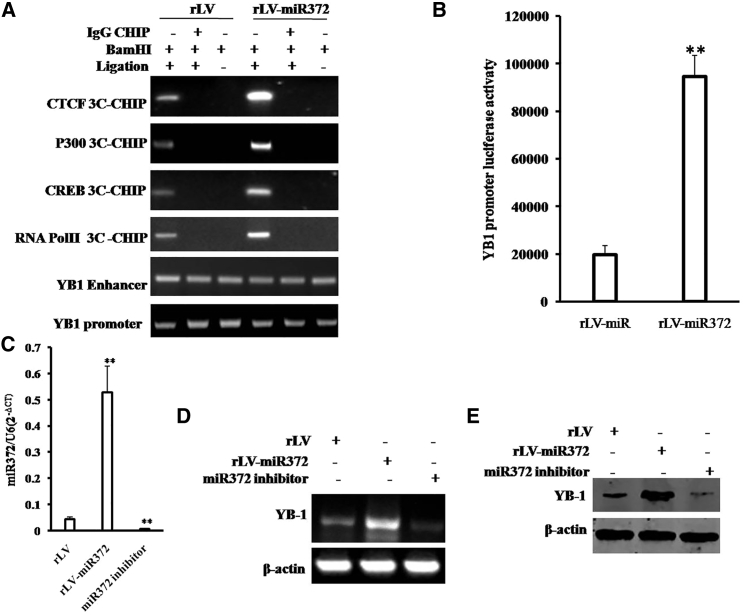
To address whether miR372 is associated with YB-1 in liver cancer, we analyzed YB-1 expression in miR372-overexpressing Hep3B cells. As shown in Figure 3A, chromosome conformation capture (3C)-chromatin immunoprecipitation (ChIP) with anti-RNA polymerase II (Pol II), anti-CTCF, anti-P300, and anti-CREB showed that miR372 promotes the formation of CTCF-mediated promoter-enhancer DNA loop of YB-1 and triggers CREB, P300, and Pol II into the DNA loop. The assay of reporter gene showed excessive miR372 enhances the YB-1 promoter luciferase activity (19,771 ± 3,843.58 versus 94,530.7 ± 8,913.75; p = 0.00474 < 0.01; Figure 3B). Ultimately, compared to rLV group, YB-1 mRNA was significantly increased in miR372-overexpressing Hep3B and decreased in Hep3B cells treated with miR372 inhibitor (Figures 3C and 3D). On the other hand, YB-1 protein is significantly increased in miR372-overexpressing Hep3B and decreased in Hep3B cells treated with miR372 inhibitor (Figure 3E). Together, these observations suggest that miR372 promotes the expression YB-1 in liver cancer cells.
Figure 3.
miR372 Enhances Expression of YB-1 in Liver Cancer Cells
(A) Chromosome conformation capture (3C)-chromatin immunoprecipitation (ChIP) with anti-RNA polymerase II, anti-CTCF, anti-P300, and anti-CREB in liver cancer cells Hep3B cell lines infected with rLV and rLV-miR372, respectively. IgG 3C-CHIP as negative control is shown. The PCR analysis is applied for detecting YB-1 promoter-enhancer coupling product using YB-1 promoter and enhancer primers. The YB-1 promoter and enhancer as INPUT are shown. (B) The assay of YB-1 promoter luciferase activity is shown. Each value was presented as mean ± SEM. **p < 0.01. (C) miR372 analysis using real-time RT-PCR and U6 as internal control is shown. (D) RT-PCR analysis with YB1 primers in liver cancer cells Hep3B cell lines infected with rLV and rLV-miR372 and treated with miR372 inhibitor, respectively, is shown. β-actin as internal control is shown. (E) Western blotting analysis with anti-YB1 in liver cancer cells Hep3B cell lines infected with rLV and rLV-miR372 and treated with miR372 inhibitor, respectively. β-actin as internal control is shown.
miR372 Enhances Phosphorylation of YB-1 by Increasing HULC and Decreasing PTEN
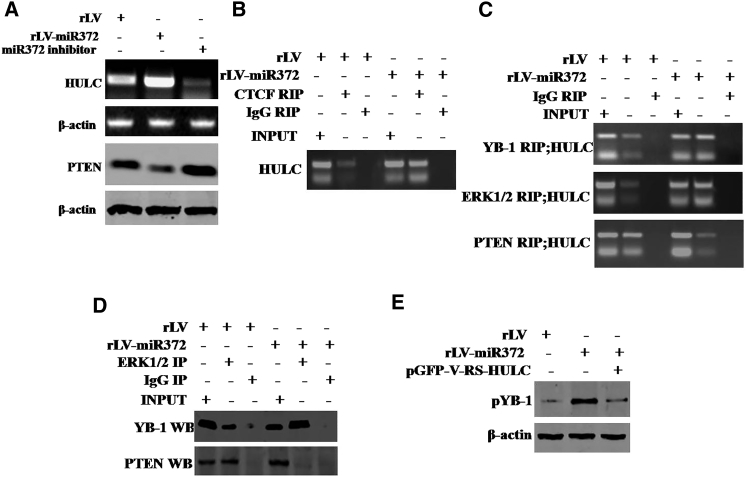
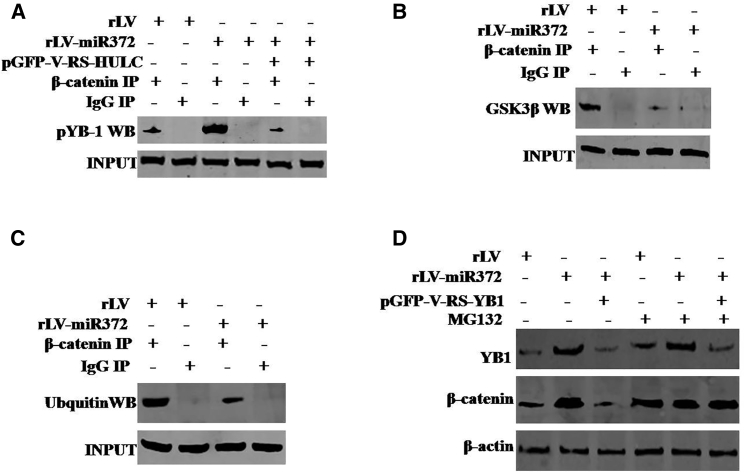
Given miR372 enhanced the expression of YB1, we wonder whether miR372 is involved in phosphorylation of YB-1. To address whether miR372 may alter phosphorylation of YB-1 dependent on noncoding RNA HULC, we first consider whether miR372 influences on HULC in liver cancer cells. As shown in Figure 4A, compared to rLV, noncoding RNA HULC was significantly increased and the expression of PTEN was significantly decreased in rLV-miR372 group. Furthermore, compared to control, excessive miR372 increased HULC looping mediated by CTCF (Figure 4B). Moreover, excessive miR372 increased the YB-1 and ERK1/2 in the HULC looping. However, the PTEN was decreased in the HULC looping in rLV-miR372 group (Figure 4C). Thereby, compared to rLV group, the interplay between YB-1 and ERK1/2 was significantly increased and the interplay between PTEN and ERK1/2 was significantly decreased in rLV-miR372 (Figure 4D). Ultimately, excessive miR372 significantly increased the phosphorylation of YB-1. However, HULC knockdown or PTEN overexpression abrogated this miR372 action (Figure 4E). Taken together, our observations suggest that miR372 enhances the phosphorylation of YB-1 in human liver cancer cells Hep3B.
Figure 4.
miR372 Enhances Phosphorylation of YB-1 in Liver Cancer Cells
(A) RT-PCR analysis with HULC primers and western blotting with anti-PTEN in liver cancer cells Hep3B cell lines infected with rLV and rLV-miR372 and treated with miR372 inhibitor, respectively. β-actin as internal control is shown. (B) RNA immunoprecipitation (RIP) anti-CTCF followed by RT-PCR with HULC primers in liver cancer cells Hep3B cell lines infected with rLV and rLV-miR372, respectively, is shown. IgG RIP as negative control is shown. HULC as INPUT is shown. (C) RIP anti-YB-1, anti-ERK1/2, and anti-PTEN followed by RT-PCR with HULC primers in liver cancer cells Hep3B cell lines infected with rLV and rLV-miR372, respectively, are shown. IgG RIP as negative control is shown. HULC as INPUT is shown. (D) Co-immunoprecipitation (coIP) with anti-ERK1 followed by western blotting with anti-YB-1 and anti-PTEN in the Hep3B cell lines infected with rLV and rLV-miR372, respectively, is shown. IgG IP as negative control is shown. INPUT refers to western blotting with anti-YB-1 and anti-ERK1/2. (E) Western blotting analysis with anti-pYB1 in liver cancer cells Hep3B cell lines infected with rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-HULC, respectively, is shown. β-actin as internal control is shown.
miR372 Inhibits the Degradation of β-Catenin Dependent on Phosphorylation of YB-1
To validate whether miR372 could regulate the expression of β-catenin dependent on phosphorylation of YB-1, we analyzed the ubiquitin modification of β-catenin in three groups, including rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-YB1 (YB1 RNAi). As shown in Figure 5A, the excessive miR372 enhances the interaction between β-catenin and pYB-1 compared to control. However, HULC knockdown completely abolished this action of miR372. In particular, the interaction between β-catenin and glycogen synthase kinase-3β (GSK3β) was weakened in rLV-miR372 group compared to rLV control group (Figure 5B). Moreover, compared to rLV control, excessive miR372 inhibited the ubiquitin modification of β-catenin (Figure 5C). Ultimately, compared to rLV group, the expression of β-catenin was significantly increased in rLV-miR372 group and the expression of β-catenin was not significantly altered in rLV-miR372 plus pGFP-V-RS-YB1. However, cells were incubated with 50 μM MG132, and the expression of β-catenin was not significantly different in these three groups of Hep3B cells infected with rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-YB-1, respectively (Figure 5D). Taken together, these observations suggest miR372 inhibits the degradation of β-catenin dependent on phosphorylation of YB-1.
Figure 5.
miR372 Inhibits the Degradation of β-Catenin Dependent on Phosphorylation of YB-1 in Liver Cancer Cells
(A) coIP with anti-β-catenin followed by western blotting with anti-pYB-1 in the Hep3B cell lines infected with rLV and rLV-miR372, respectively. IgG IP as negative control is shown. INPUT refers to western blotting with anti-YB-1 and anti-β-catenin. (B) coIP with anti-β-catenin followed by western blotting with anti-GSK3β in the Hep3B cell lines infected with rLV and rLV-miR372, respectively, is shown. IgG IP as negative control is shown. INPUT refers to western blotting with anti-GSK3β and anti-β-catenin. (C) coIP with anti-β-catenin followed by western blotting with anti-ubiquitin in the Hep3B cell lines infected with rLV and rLV-miR372, respectively, is shown. IgG IP as negative control is shown. INPUT refers to western blotting with ubiquitin and anti-β-catenin. (D) Cells were incubated with 50 μM MG132 (Sigma) for 6 hr after infection and western blotting analysis with anti-β-catenin in liver cancer cell line Hep3B cells infected with rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-YB-1, respectively. β-actin as internal control is shown.
miR372 Enhances the Expression and Activity PKM2 by Activating β-Catenin
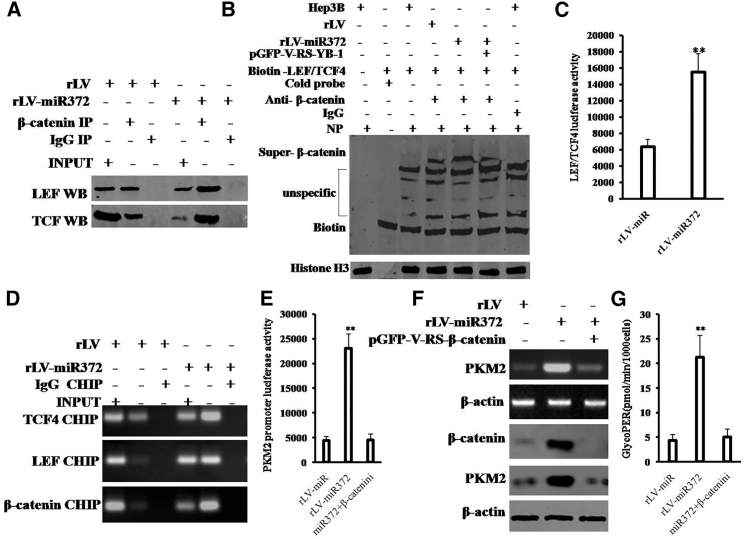
Given that miR372 enhanced the expression of β-catenin dependent on pYB-1, we wonder whether miR372 could influence on the expression and activity PKM2 through activating β-catenin in human liver cancer cells. As shown in Figure 6A, compared to rLV group, the interaction between β-catenin and LEF was significantly increased and the interaction between β-catenin and TCF4 was significantly increased in rLV-miR372 group. Moreover, Super-electrophoretic mobility shift assay (EMSA) (gel-shift) with biotin-LEF/TCF4 probe showed that excessive miR372 enhanced the binding of LEF/TCF4 probe to β-catenin compared to control. However, YB-1 knockdown fully abrogated this action of miR372 (Figure 6B). Furthermore, LEF/TCF4 luciferase activity was significantly increased in rLV-miR372 group compared to the rLV group (6,381.3 ± 858.57 versus 15,542 ± 2,222.07; p = 0.00397 < 0.01; Figure 6C). Strikingly, the loading of LEF or TCF4 on PKM2 promoter region was significantly increased in rLV-miR372 group compared to the rLV group (Figure 6D). And the PKM2 promoter luciferase activity was significantly increased in rLV-miR372 group compared to the rLV group (4,416.67 ± 811.46 versus 23,032.3 ± 2974; p = 0.0022 < 0.01). However, β-catenin knockdown fully abrogated this action of miR372 (4,416.67 ± 811.46 versus 4,553 ± 1,169.5; p = 0.4563 > 0.05; Figure 6E). Furthermore, as shown in Figure 6F, both PKM2 mRNA and PKM2 protein significantly increased in rLV-miR372 group compared to the rLV group (p < 0.01). However, β-catenin knockdown fully abrogates this function of miR372. In particular, glycolytic proton efflux rate (glycoPER) was significantly increased in rLV-miR372 group compared to the rLV group (4.31 ± 1.27 versus 21.22 ± 4.49; p = 0.0061 < 0.01). However, β-catenin knockdown fully abrogates this function of miR372 (4.31 ± 1.27 versus 5.06 ± 1.63; p = 0.26551 > 0.05; Figure 6G). Together, these observations suggest that miR372 enhances the expression and activity of PKM2 by activating β-catenin.
Figure 6.
miR372 Enhances the PKM2 by Enhancing the Activation of β-Catenin in Liver Cancer Cells
(A) coIP with anti-β-catenin followed by western blotting with anti-LEF and anti-TCF4 in the Hep3B cell lines infected with rLV and rLV-miR372, respectively. IgG IP as negative control is shown. INPUT refers to western blotting with anti-TCF4 and anti-LEF. (B) Super-EMSA (gel-shift) with biotin-LEF/TCF4 probe and anti-β-catenin antibody is shown. The intensity of the band was examined by western blotting with anti-biotin. Histone H3 as internal control is shown. (C) The assay of LEF/TCF4 luciferase activity is shown. Each value was presented as mean ± SEM. **p < 0.01. (D) ChIP with anti-LEF and anti-TCF4 followed by PCR with PKM2 promoter primers in the Hep3B cell lines infected with rLV and rLV-miR372, respectively, is shown. IgG CHIP as negative control is shown. PKM2 promoter as INPUT is shown. (E) The assay of PKM2 promoter luciferase activity is shown. Each value was presented as mean ± SEM. **p < 0.01. (F) Western blotting analysis with anti-PKM2 and RT-PCR with PKM2 primers in liver cancer cells Hep3B cell lines infected with rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-β-catenin, respectively, is shown. β-actin as internal control is shown. (G) The assay of glycolytic proton efflux rate (glycoPER) in liver cancer cells Hep3B cell lines infected with rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-β-catenin, respectively, is shown.
miR372 Promotes the Expression of erbB-2 Dependent on PKM2 Activity
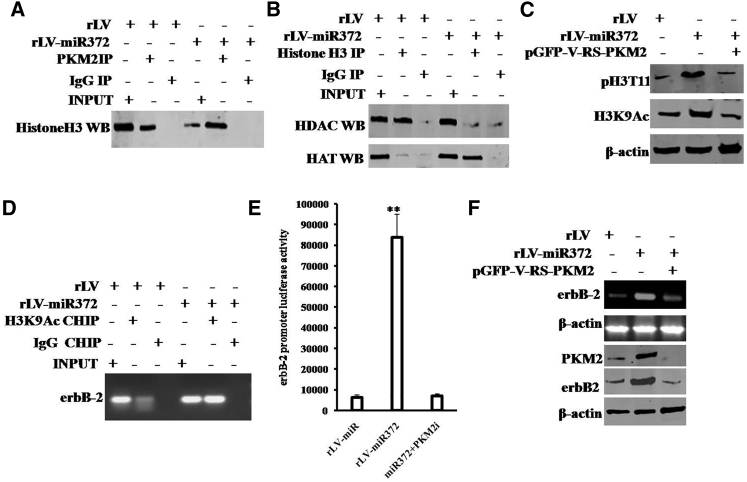
It is well known that erbB-2 is an oncogene and highly expressed in several cancers. To validate whether miR372 could influenced on the expression of erbB-2 dependent on PKM2 activity in liver cancer, we performed related experiments in the Hep3B cell lines infected with rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-PKM2 (PKM2 RNAi). As shown in Figure 7A, the interaction between PKM2 and histone H3 was significantly increased in rLV-miR372 group compared to rLV group. Moreover, compared to rLV group, the interaction between HDAC and histone H3 was significantly decreased, and the interaction between HAT and histone H3 was significantly increased in rLV-miR372 group (Figure 7B). In particular, the expression of both pH3T11 and acetylation on histone H3 lysine 9 (H3K9Ac) was significantly increased in rLV-miR372 group compared to rLV group. However, the expression of neither pH3T11 nor H3K9Ac was significantly altered in liver cancer cells Hep3B cell lines infected with rLV-miR372 plus pGFP-V-RS-PKM2 compared to rLV group (Figure 7C). Furthermore, the loading of H3K9Ac on the promoter region of erbB-2 was significantly increased in rLV-miR372 group compared to rLV group (Figure 7D). Furthermore, the erbB-2 promoter luciferase activity was significantly increased in rLV-miR372 group compared to rLV group (6,269.33 ± 1,317.609 versus 83,809 ± 11,185.22; p = 0.00029 < 0.01). However, PKM2 knockdown fully abrogates this action of miR372 (6,269.33 ± 1,317.609 versus 6,942 ± 1,060.67; p = 0.2488 > 0.05; Figure 7E). Finally, both the erbB-2 mRNA and erbB-2 protein were significantly increased in rLV-miR372 group compared to rLV group. However, the expression of neither erbB-2 mRNA nor erbB-2 protein was significantly altered in Hep3B cells infected with rLV-miR372 plus pGFP-V-RS-PKM2 compared to rLV group (Figure 7F). Together, the observations indicate that miR372 enhances the transcription and translation of erbB-2 dependent on PKM2 activity in human liver cancer cells.
Figure 7.
miR372 Promotes the erbB-2 through PKM2 in Liver Cancer Cells
(A) coIP with anti-PKM2 followed by western blotting with anti-histone H3 in the Hep3B cell lines infected with rLV and rLV-miR372, respectively. IgG IP as negative control is shown. INPUT refers to western blotting with anti-PKM2 and anti-histone H3. (B) coIP with anti-DHAC3 followed by western blotting with anti-histone H3 in the Hep3B cell lines infected with rLV and rLV-miR372, respectively, is shown. IgG IP as negative control is shown. INPUT refers to western blotting with anti-DHAC3 and anti-histone H3. (C) Western blotting analysis with anti-pH3T11 and anti-H3K9Ac in liver cancer cells Hep3B cell lines infected with rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-PKM2, respectively, is shown. β-actin as internal control is shown. (D) ChIP with anti-H3K9Ac followed by PCR with ErbB-2 promoter primers in the Hep3B cell lines infected with rLV and rLV-miR372, respectively, is shown. IgG CHIP as negative control is shown. PKM2 promoter as INPUT is shown. (E) The assay of ErbB-2 promoter luciferase activity is shown. Each value was presented as mean ± SEM. **p < 0.01. (F) Western blotting analysis with anti-ErbB-2 and RT-PCR with erbB-2 primers in liver cancer cells Hep3B cell lines infected with rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-PKM2, respectively, is shown. β-actin as internal control is shown.
Both erbB-2 and YB-1 Decide on the Oncogenic Function of miR372
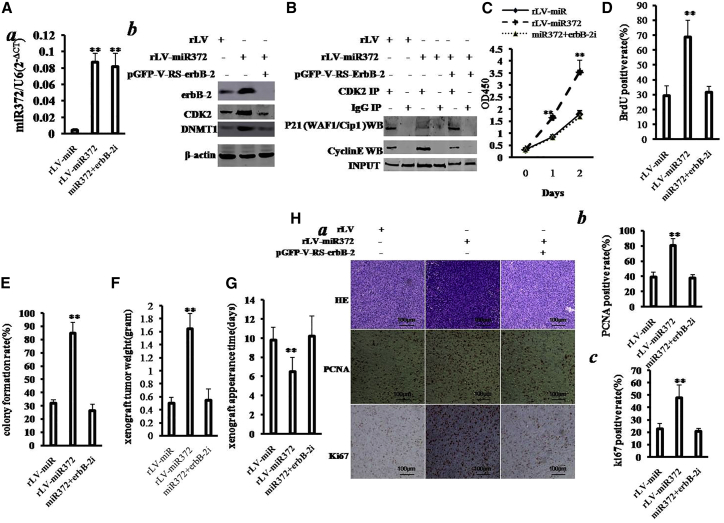
Indeed, miR372 enhances the expression of erbB-2 through PTEN-CTCF-HULC-pYB1-β-catenin-PMK2 pathway. To identify whether miR372 oncogenic function is associated with erbB-2, we performed the rescued experiments in the three stable Hep3B lines infected with rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-erbB-2 (erbB-2 RNAi). As shown in Figure 8Aa, compared to rLV group, miR372 was overexpressed in groups of rLV-miR372 and rLV-miR372 plus pGFP-V-RS-erbB-2, respectively. And shown in Figure 8Ab, erbB-2 was increased in groups of rLV-miR372 and decreased in rLV-miR372 plus pGFP-V-RS-erbB-2 group compared to rLV group. Both CDK2 and DNMT1 were increased in groups of rLV-miR372; however, neither CDK2 nor DNMT1 is significantly altered in rLV-miR372 plus pGFP-V-RS-ErbB-2 group. Next, we performed the co-immunoprecipitation (coIP) with anti-CDK2 in these cell lines. As shown in Figure 8B, the interaction between CDK2 and cyclin E is significantly increased in rLV-miR372 group compared to rLV group, and the interaction between CDK2 and P21/WAF1/Cip1 was significantly decreased in rLV-miR372 group compared to rLV group. However, erbB-2 knockdown fully abrogates this function of miR372 in Hep3B cells.
Figure 8.
Rescued erbB-2 Abrogates Oncogenic Action of miR372
(A) Western blotting analysis with anti-erbB-2, anti-CDK2, and anti-DNMT1 in liver cancer cell line Hep3B cells infected with rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-erbB-2, respectively. β-actin as internal control is shown. (B) coIP with anti-CDK2 followed by western blotting with anti-cyclin E in the Hep3B cells infected with rLV, rLV-miR372, and rLV-miR372 plus pGFP-V-RS-erbB-2, respectively, is shown. IgG IP as negative control is shown. INPUT refers to western blotting with anti-CDK2. (C) Proliferation assay was performed using the CCK8. Data are means of value from three independent experiments; mean ± SEM. **p < 0.01; *p < 0.05. (D) Cell BrdU assay is shown. Data are means of value from three independent experiments, mean ± SEM. **p < 0.01; *p < 0.05. (E) (Upper) The photography of colonies from the cell lines indicated in left is shown. (Lower) Cell plate colony formation ability assay is shown. Data are means of value from three independent experiments, mean ± SEM. **p < 0.01; *p < 0.05. (F) The xenograft tumors weight (gram) in the three groups is shown. Data were means of value from six BALB/c mice; mean ± SEM; n = 6; *p < 0.05; **p < 0.01. (G) The xenograft tumors appearance time (days) in the three groups is shown. Data were means of value from six BALB/c mice; mean ± SEM; n = 6; *p < 0.05; **p < 0.01. (H) (a) A portion of each xenograft tumor was fixed in 4% formaldehyde and embedded in paraffin, and the micrometers of sections (4 μm) were made for H&E staining and PCNA staining (original magnification ×100). (b) PCNA and Ki67 positive rate is shown.
Then, we detected the ability of cell proliferation. As shown in Figure 8C, excessive miR372 significantly increased the growth of liver cancer cell Hep3B compared to the control cells (p < 0.01). However, excessive miR372 plus erbB-2 knockdown could significantly not alter the growth ability of liver cancer cells compared to rLV group (p > 0.05). Moreover, excessive miR372 significantly increased the BrdU-positive rate compared to the control cells (29.07% ± 6.96% versus 68.66% ± 11.47%; p = 0.013 < 0.05). However, excessive miR372 plus ErbB-2 knockdown could not significantly alter the BrdU-positive rate of liver cancer cells (29.07% ± 6.96% versus 31.54% ± 4.23%; p = 0.335 > 0.05; Figure 8D).We Further performed colony formation assay and observed a significant increase in colony formation efficiency rate in excessive miR372 group (31.82% ± 2.63% versus 84.72% ± 8.26%; p = 0.006532 < 0.01). However, excessive miR372 plus erbB-2 knockdown could not significantly alter the colony formation rate of liver cancer cells (31.82% ± 2.63% versus 26.43% ± 4.70%; p = 0.162 > 0.05; Figure 8E). Furthermore, the three stable Hep3B cell lines were injected subcutaneously into athymic Balb/C mice. As shown in Figure 8F, when miR372 was overexpressed, the average xenograft tumor weight increased approximately three-fold compared to the corresponding control group (0.497 ± 0.092 g versus 1.647 ± 0.2365 g; p = 0.0012 < 0.01). However, excessive miR372 plus erbB-2 knockdown could not significantly alter the xenograft tumor weight (0.497 ± 0.092 g versus 0.545 ± 0.171 g; p = 0.2419 > 0.05). On the other hand, when miR372 was overexpressed, the average xenograft tumor appearance time was decreased compared to the corresponding control group (9.83 ± 1.32 days versus 6.5 ± 1.52 days; p = 0.0044 < 0.01). However, excessive miR372 plus erbB-2 knockdown could significantly not alter the xenograft tumor appearance (9.83 ± 1.32 days versus 10.12 ± 2.14 days; p = 0.371 > 0.05; Figure 8G). Moreover, as shown in Figures 8Ha and 8Hb, the expression of PCNA was significantly higher in miR372-overexpressing xenograft tumors compared to the control group (39.23% ± 6.61% versus 80.4% ± 9.15%; p = 0.0089 < 0.01). However, excessive miR372 plus erbB-2 knockdown could significantly not alter the PCNA positive rate (39.23% ± 6.61% versus 37.73% ± 4.86%; p = 0.157 > 0.05). Moreover, the expression of Ki67 was significantly higher in miR372-overexpressing xenograft tumors compared to the control group (22.7% ± 4.59% versus 47.77% ± 10.39%; p = 0.008704 < 0.01). However, excessive miR372 plus erbB-2 knockdown could not significantly alter the Ki67 positive rate (22.7% ± 4.59% versus 20.57% ± 2.57%; p = 0.1386 > 0.05).
Furthermore, in order to address whether YB-1 knockdown or erbB-2 knockdown influences the functions of miR372 in human liver cancer cell line Huh7, we first constructed four stable Huh7 cell lines, including rLV, rLV-miR372, rLV-miR372 plus pGFP-V-RS-YB-1, and rLV-miR372 plus pGFP-V-RS-erbB-2, respectively. As shown in Figure S3A, mature miR372 was significantly overexpressed in rLV-miR372 group, rLV-miR372 plus pGFP-V-RS-YB-1 group, and rLV-miR372 plus pGFP-V-RS-erbB-2 group, respectively, compared to control group. As shown in Figure S3B, compared to rLV Huh7 group, the expression of YB-1 and erbB-2 is significantly increased in rLV-miR372 Huh7 group, and the expression of YB-1 and erbB-2 is significantly decreased in rLV-miR372 plus pGFP-V-RS-YB-1 Huh7 group. In particular, the expression of erbB-2 is significantly decreased in rLV-miR372 plus pGFP-V-RS-erbB-2 Huh7 group but the expression of YB-1 not. Moreover, as shown in Figure S4A, the colony formation efficiency rate is significantly increased in rLV-miR372 Huh7 group compared to control (79.6% ± 9.6% versus 34.8% ± 6.46%; p = 0.0009 < 0.01). However, the colony formation efficiency rate is not significantly altered in rLV-miR372 plus pGFP-V-RS-YB-1 Huh7 group (31.6% ± 2.86% versus 34.8% ± 6.46%; p = 0.2489 > 0.05) and rLV-miR372 plus pGFP-V-RS-erbB-2 Huh7 group, respectively (29.8% ± 4.55% versus 34.8% ± 6.46%; p = 0.079 > 0.05). As shown in Figure S4B, the xenograft tumor weight is significantly increased in rLV-miR372 Huh7 group compared to control (1.075 ± 0.255 g versus 0.602 ± 0.199 g; p = 0.005 < 0.01). However, the xenograft tumor weight is not significantly altered in rLV-miR372 plus pGFP-V-RS-YB-1 Huh7 group (0.53 ± 0.166 g versus 0.602 ± 0.199 g; p = 0.1709 > 0.05) and rLV-miR372 plus pGFP-V-RS-erbB-2 Huh7 group, respectively (0.568 ± 0.109 g versus 0.602 ± 0.199 g; p = 0.3253 > 0.05). As shown in Figure S4C, the PCNA positive rate in xenograft tumor is significantly increased in rLV-miR372 Huh7 group compared to control (77.73% ± 12.61% versus 37.13% ± 8.43%; p = 0.0005 < 0.01). However, the PCNA positive rate in xenograft tumor is significantly not altered in rLV-miR372 plus pGFP-V-RS-YB-1 Huh7 group (34.58% ± 10.12% versus 37.13% ± 8.43%; p = 0.3527 > 0.05) and rLV-miR372 plus pGFP-V-RS-erbB-2 Huh7 group, respectively (32.72% ± 3.28% versus 37.13% ± 8.43%; p = 0.1903 > 0.05).
These results suggest that miR372 promotes cell growth, colony formation ability, and cell growth in vivo. However, both YB-1 knockdown and ErbB-2 knockdown fully abrogate this function of miR372. Taken together, both erbB-2 and YB-1 determine oncogenic function of miR372 in human liver cancer cells.
Discussion
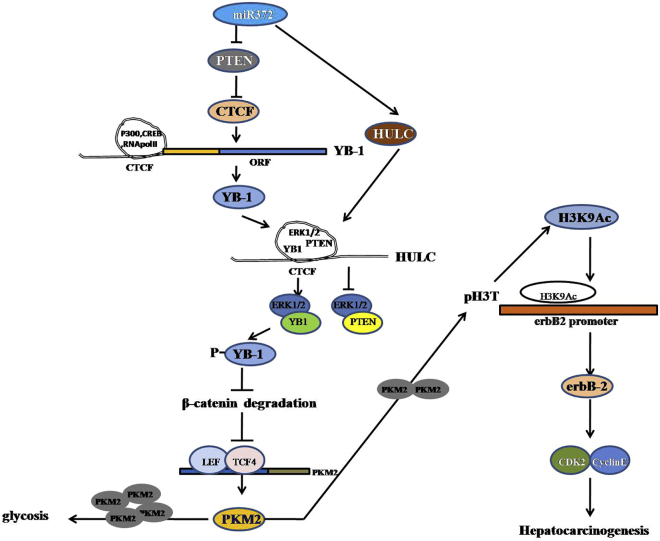
Recent studies have shown that miR-372 plays important roles in HCC progression. However, results have been conflicting regarding its expression levels and role in HCC. In the report, our findings indicated the effects of miR372 during hepatocarcinogenesis. Herein, our results demonstrate that miR372 promotes liver cancer cell cycle progress by activating cell cycle complex CDK2-cyclin E-P21/Cip1/WAF1 through miR372-YB-1-β-catenin-LEF/TCF4-PKM2-erbB-2 axis (Figure 9).
Figure 9.
The Schematic Illustrates a Model of miR372 Promotes Liver Cancer Cell Growth by Upregulating erbB-2 through Enhancing YB-1 via Targeting for PTEN
miR372 accelerates malignant progression of liver cancer cells in vitro and in vivo. Mechanistically, miR372 enhances expression of YB-1 and consequently promotes phosphorylation of YB-1 via HULC looping dependent on ERK1/2 and PTEN. Moreover, miR372 inhibits the degradation of β-catenin dependent on phosphorylation of YB-1 and then enhances the expression and activity PKM2 by β-catenin-LEF/TCF4 pathway. Furthermore, the loading of LEF/TCF4 on PKM2 promoter region is significantly increased in miR372-overexpressed liver cancer cells. Ultimately, miR372 promotes the expression of erbB-2 through PKM2-pH3T11-H3K9Ac pathway.
To date, accumulating evidence indicates that miR372 results in a critical role in carcinogenesis and cancer metastasis. For example, miR372 expression correlates with prognosis and metastasis in hepatocellular carcinoma.18 Herein, our findings demonstrate that miR372 accelerates malignant progression of liver cancer cells, including Hep3B and Huh7. These findings are noteworthy that miR372 is a key oncogene mediating various biological processes, including cell proliferation and differentiation.
Notably, our observations suggest that miR372 promotes the expression YB-1 by targeting PTEN directly. Our findings in this study provide novel evidence for an active role of miR372 in this action. This assertion is based on several observations.1 miR372 targets for PTEN to increase the expression of CTCF.2 miR372 promotes the formation of CTCF-mediated promoter-enhancer DNA loop of YB-1 and triggered CREB, P300, and Pol II into the DNA loop.3 Excessive miR372 enhanced the YB-1 promoter luciferase activity.4 YB-1 mRNA and protein was significantly increased in miR372-overexpressing Hep3B and Huh7 and decreased in Hep3B cells treated with miR372 inhibitor. It has been identified that YB1, which is overexpressed in many types of human cancers, is a multifunctional transcription factor with vital roles in proliferation, differentiation, and apoptosis.19 In particular, YB1 is also involved in regulating G2/M phase by regulating multiple cell-cycle-related genes.20 However, a report shows thatYB1 suppressed liver tumor growth and metastasis in a nude mice liver tumor model.21 Also the upregulation of YB-1 expression activates the NF-κB signaling pathway.22 Strikingly, YB-1 is related to cancer therapy. For example, human helicase RECQL4 drives cisplatin resistance in gastric cancer by activating an AKT-YB1-MDR1 signaling pathway23 and the SHH:YAP:YB1:IGF2 axis as a powerful target for therapeutic intervention in medulloblastomas.24 Also,YB-1 associates with thousands of nonpolyadenylated short RNAs (shyRNAs) that are further processed into small RNAs (smyRNAs).25 Moreover, ENZ-induced HER2 overexpression was dependent on AKT-YB1 activation and modulated AR activity.26, 27 Intrguingly,YB1/p32 interacts with Msx1 homeoprotein and functions as a regulator of C2C12 myoblast differentiation.28 Thereby, YB-1 may decide the function of miR372 in hepatocarcinogenesis.
Furthermore, our observations demonstrate that miR372 enhances phosphorylation of YB-1 via increasing HULC and decreasing PTEN. Our findings in this study provide novel evidence for an active role of miR372 in this action. This assertion is based on several observations. (1) Noncoding RNA HULC is significantly increased and the expression of PTEN is significantly decreased in rLV-GLP2-infected Hep3B cells. (2) Excessive miR372 increases HULC looping mediated by CTCF. (3) Excessive miR372 increases the YB-1 and ERK1/2 in HULC looping and decreases the PTEN in HULC looping. (4) Excessive miR372 significantly increases the interplay between YB-1 and ERK1/2 and between PTEN and ERK1/2. (5) Excessive miR372 significantly increases the phosphorylation of YB-1. In particular, HULC knockdown or PTEN overexpression abrogates the functions of miR372. A study shows that long non-coding RNA (lncRNA) HULC (highly upregulated in liver cancer) is involved in autophagy and chemoresistance of HCC29 and modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase to enhance hepatocarcinogenesis.30 Also, miR-372 has been shown to play a role in the regulation of HULC and PTEN through a competing endogenous RNA (ceRNA) mechanism.31 Furthermore, the PTEN gene, an important tumor-suppressor gene, has been demonstrated to have the potential for inhibiting proliferation, migration, and invasion in various types of cancer cells through PTEN/Akt/mTOR pathway.32, 33 Also, Plk1 phosphorylation of PTEN promotes tumorigenesis in both its phosphatase-dependent and independent pathways.34 Moreover, a report shows that transforming growth factor β1 (TGF-β1) stimulates migration of type II endometrial cancer cells by downregulating PTEN via activation of SMAD and ERK1/2 signaling pathways.35 In addition, MeCP2 facilitates gastric cancer cell proliferation through activation of the MEK1/2-ERK1/2 signaling pathway by upregulating GIT1.36
Strikingly, our data suggest that miR372 inhibits the degradation of β-catenin dependent on phosphorylation of YB-1.This assertion is identified by several observations. (1) The excessive miR372 enhances the interaction between β-catenin and pYB-1; however, HULC knockdown completely abolished this action of miR372. (2) The interaction between β-catenin and GSK3β was weaken in rLV-miR372 group compared to rLV group. (3) Excessive miR372 inhibited the ubiquitin modification of β-catenin. (4) Ultimately, the expression of β-catenin was significantly increased in rLV-miR372 group compared to rLV group; however, the expression of β-catenin was significantly not altered in rLV-miR372 plus pGFP-V-RS-YB1 miR372 compared to control. A report shows that active canonical Wnt signaling results in recruitment of β-catenin to DNA by TCF/LEF family members, leading to transcriptional activation of TCF target genes.37 And silencing of Glut1 induces chemoresistance via modulation of Akt/GSK-3β/β-catenin/survivin signaling pathway in breast cancer cells.38 Also, diaphanous-related formin 3 (DIAPH3) promotes the growth, migration, and metastasis of HCC by activating beta-catenin/TCF signaling.39 Furthermore, GSK-3β suppresses cancer by inhibiting Wnt/β-catenin signaling pathway40, 41 and PI3K/Akt survival pathway.42
Moreover, our results indicate that miR372 enhances the expression and activity of PKM2 by β-catenin; the functions of miR372 promotion of liver cancer cell growth are explained by results from seven parallel sets of experiments. (1) The interaction between β-catenin and LEF was significantly increased, and the interaction between β-catenin and TCF4 was significantly increased in rLV-miR372 group compared to rLV group. (2) Excessive miR372 enhanced the binding of LEF/TCF4 probe to β-catenin compared to control. However,YB-1 knockdown fully abrogates this action of miR372. (3) LEF/TCF4 luciferase activity was significantly increased in rLV-miR372 group compared to the rLV group. (4) The loading of LEF or TCF4 on PKM2 promoter region was significantly increased in rLV-miR372 group compared to the rLV group. (5) The PKM2 promoter luciferase activity was significantly increased; however, β-catenin knockdown fully abrogated the action of miR372. (6) Both PKM2 mRNA and PKM2 protein significantly increased in rLV-miR372 group compared to the rLV group; however, β-catenin knockdown abrogated the functions of miR372. (7) GlycoPER is significantly increased in rLV-miR372 group compared to the rLV group; however, β-catenin knockdown abrogates the functions of miR372. It is well known that PKM2 catalyzes the last step of glycolysis and plays an important role in tumor cell proliferation. Recent studies have reported that PKM2 also regulates apoptosis by stabilizing Bcl2.43 PKM2 is a key kinase of glycolysis and is characteristic of all proliferating cells and promotes cell migration and inhibits autophagy by mediating phosphatidylinositol 3-kinase (PI3K)/AKT activation and contributes to the malignant development of gastric cancer.44 Moreover, small molecule activators of PKM2 suppress tumor formation.45 On the other hand, PKM2 has been thought to favor cell proliferation46 and differentiation.47, 48
Strikingly, our results indicated that miR372 promotes the erbB-2 through PKM2 epigenetically. Our findings in this study provide novel evidence for an active role of miR372 in this action. This assertion is based on several observations. (1) The interaction between PKM2 and histone H3 was significantly increased in rLV-miR372 group compared to rLV group. (2) The interaction between DHAC and histone H3 was significantly decreased and the interaction between HAT and histone H3 was significantly increased in rLV-miR372 group compared to rLV group. (3) The expression of both pH3T11 and H3K9Ac was significantly increased in rLV-miR372 group compared to rLV group. However, the expression of neither pH3T11 nor H3K9Ac is significantly altered in liver cancer cells Hep3B cell lines infected with rLV-miR372 plus pGFP-V-RS-PKM2 compared to rLV group. (4) The loading of H3K9Ac on the promoter region of erbB-2 is significantly increased in rLV-miR372 group compared to rLV group. (5) The erbB-2 promoter luciferase activity is significantly increased in rLV-miR372 group; however, PKM2 knockdown fully abrogated the action of miR372. (6) Both erbB-2 mRNA and protein are significantly increased in rLV-miR372 group compared to rLV group. However, the expression of neither erbB-2 mRNA nor erbB-2 protein is significantly altered in liver cancer cells Hep3B cell lines infected with rLV-miR372 plus pGFP-V-RS-PKM2 compared to rLV group. A study indicates that H3K9Ac mediates a switch from transcription initiation to elongation.49 Furthermore, histone modifiers and marks define heterogeneous groups of colorectal carcinomas and affect responses to histone deacetylase (HDAC) inhibitors in vitro.50 Recent studies showed that the rate-limiting glycolytic enzyme, PKM2, directly phosphorylates H3 at threonine 11 (H3T11) to regulate gene expression and cell proliferation and glycolysis regulates H3T11 phosphorylation by fueling the substrate, phosphoenonylpyruvate, and the coactivator, FBP, to Pyk1.51
Importantly, our results indicated that erbB-2 knockdown abrogates oncogenic action of miR372. This assertion is based on several observations. (1) The interaction between CDK2 and cyclin E was significantly increased in rLV-miR372 group compared to rLV group, and the interaction between CDK2 and P21/WAF1/Cip1 was significantly decreased in rLV-miR372 group compared to rLV group. However, erbB-2 knockdown fully abrogated this miR372 function. (2) Excessive miR372 significantly increased the growth of liver cancer cell compared to the control cells. However, excessive miR372 plus erbB-2 knockdown or plus YB-1 knockdown could not significantly alter the growth ability of liver cancer cells compared to rLV group. Moreover, excessive miR372 significantly increased the BrdU positive rate compared to the control cells. However, excessive miR372 plus erbB-2 knockdown or plus YB-1 knockdown could not significantly alter the BrdU positive rate of liver cancer cells. (3) Colony formation efficiency rate is significantly increased in excessive miR372 group; however, excessive miR372 plus erbB-2 knockdown or plus YB-1 knockdown could significantly not alter the colony formation rate of liver cancer cells. (4) When miR372 is overexpressed, the average xenograft tumor weight increased approximately three-fold compared to the corresponding control group; however, excessive miR372 plus erbB-2 or plus YB-1 knockdown could not significantly alter the xenograft tumor weight. It is well known that the erbB family is unique among the various groups of receptor tyrosine kinases (RTKs) in that erbB3 has impaired kinase activity, whereas erbB-2 does not have a direct ligand. The overexpression and over-activation of erbB receptors are correlated with poor prognosis, drug resistance, cancer metastasis, and lower survival rate. erbB receptors, especially epidermal growth factor receptor (EGFR) and erbB-2, have been the primary choices as targets for developing cancer therapies.52 erbB-2-positive mammary tumors can escape PI3K-p110α loss through downregulation of the PTEN tumor suppressor.53 Moreover, erbB-2 signaling epigenetically suppresses miR-205 transcription via the Ras/Raf/MEK/ERK pathway,54 and HCaRG/COMMD5 inhibits erbB-receptor-driven renal cell carcinoma.55 Moreover, ERRF sensitizes erbB-2-positive breast cancer cells to lapatinib treatment likely by attenuating MCL1 and ERBB2 expression.56 In addition, overexpression of erbB-2 has been implicated in the pathogenesis of cholangiocarcinoma, suggesting that combined erbB-2 targeting might serve as a target-based therapeutic strategy for this highly lethal cancer.57 In addition, a report suggests that erbB-2/beta-catenin upregulation contributes importantly to the mechanism of HBxAg-mediated hepatocellular carcinoma growth.58
In the present study, we clearly indicate that miR372 inhibits the interaction between P21 (WAF1/Cip1) and CDK2 and increases the interaction between CDK2 and cyclin E and enhanced the expression of DNMT1. It is well known P21 (CiP1/WAF1) acts a suppressor in the early stage of cancer invasiveness.59 Inhibition of p21 (WAF1/CIP1) promotes carcinogenesis.60 Furthermore, lower p21WAF1/cip1 is associated with cancer recurrence and poor prognosis.61, 62 The p21 effect on the cells to response to gefitinib was further confirmed by p21 overexpression and knockdown studies, pointing to a requirement of p21 for the cells sensitive to gefitinib.63 Furthermore, our present results show that miR372 oncogenic action is associated with CDK2 and cyclin E activity. It is well known that CDK2 regulates and controls cell cycle progression through several signaling pathways, e.g., EGF-ELK4/c-Fos pathway.64 Of significance, specific cyclin E/CDK2 inhibitors can block tumor progression.65 The G(0)/G(1) arrest was attributed to a decreased expression of cyclin D1, cyclin E, CDK2, and CDK6, especially the upregulation of p21.66 In addition, our results show that miR372 enhances the expression of DNMT1, consistent with some reports, e.g., TGF-β-mediated repression of MST1 by DNMT1 promotes glioma malignancy67 and lncRNA SNHG1 promotes DNMT1 expression, which facilitates the gastric cancer proliferation.68
Conclusions
The present study indicates a novel evidence for miR372 in hepatocarcinogenesis, which may have potential therapeutic significance. Alteration of the expression of miR372 may also mediate changes to affect gene expression and contribute to hepatocarcinogenesis. Blocking miR372 might represent a promising treatment strategy targeting tumors.
Materials and Methods
Cell Lines and Plasmids
Human hepatoma cell lines Hep3B and Huh7 were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). These cell lines were maintained in DMEM (Gibco BRL Life Technologies) supplemented with 10% fetal bovine serum (Gibco BRL Life Technologies). Lentivirus rLV-miR and rLV-miR372 were purchased from Wu Han Viral Therapy Technologies. pGFP-V-RS, pGFP-V-RS-YB1, pGFP-V-RS-β-catenin, and pGFP-V-RS-erbB-2 were purchased from Origene (Rockville, MD, USA). pGFP-V-RS-HULC was prepared by our lab.
RT-PCR
Total RNA was purified using Trizol (Invitrogen) according to manufacturer’s instructions. Primers include as follows: PKM2 promoter (P1: 5′-CAGATGCCCAGCTCTGCGCT-3′; P2: 5′-GAGCGGCAGTAGGGAGAACT-3′); PKM2 mRNA (P1: 5′-TCCTGGAGCAcATGTGCCGC-3′; P2: 5′-CCCAAACTTCAGATCCTGGA-3′); YB1 mRNA (P1: 5′-GAGCAGCGAGGCCGAGACCC-3′; P2: 5′-CTCCTGCACCCTGGTTGTCA-3′); YB1 promoter (P1: 5′-AAAATATTAGCCAATAGAAG-3′; P2: 5′-GCTGCGGCAGCTGCGGCTCC-3′); YB1 enhancer (P1: 5′-AAACTTATCTCCCTTCATGT; P2: 5′-GGTCTCTCCCGTTTTAAAAT-3′); YB1 (3C-CHIP) (P1: 5′-AAAATATTAGCCAATAGAAG-3′; P2: 5′-GGTCTCTCCCGTTTTAAAAT-3′); erbB-2 mRNA (P1: 5′-GAACCAGCCAGATGTTCGGC-3′; P2: 5′-GTTCTCTGCCGTAGGTGTCC-3′); β-actin (P1: 5′-CTTCCTTCCTGGGCATGGAG-3′; P2: 5′-TGGAGGGGCCGGACTGGTCA-3′); and HULC (P1: 5′-AACCTCCAGAACTGTGAT-3′; P2: 5′-CATAATTCAGGGAGAAAG-3′).
MicroRNA Detection
Total RNA was isolated from cultured cells using Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Real-time RT-PCR-based detection of mature miR372 and U6 small nuclear RNA (snRNA) was achieved with the miRNA Detection kit (including a universe primer and U6 primers; QIAGEN) and miR372-specific upstream primers (5′-CCTCAAATGTGGAGCACTATTCT-3′).
coIP
Cell protein is precleared with 30 μL protein G/A-plus agarose beads (Santa Cruz Biotechnology, CA) for 1 hr at 4°C. Precleared homogenates were incubated with antibody and/or normal mouse/rabbit immunoglobulin G (IgG) with rotation for 4 hr at 4°C. The immunoprecipitates were incubated with 30 μL protein G/A-plus agarose beads by rotation overnight at 4°C and then centrifuged at 3,000 rpm for 5 min at 4°C. The precipitates are washed five times for 10 min with beads wash solution. Western blotting was performed with related antibodies.
RNA Immunoprecipitation
Ribonucleoprotein-particle-enriched lysates are incubated with protein A/G-plus agarose beads (Santa Cruz Biotechnology, CA) together with the antibody or normal mouse or rabbit IgG for 4 hr at 4°C. Beads were subsequently washed four times with 50 mM Tris-HCl (pH 7.0), 150 mM NaCl, 1 mM MgCl2, and 0.05% NP-40 and twice after addition of 1M urea. Total RNA is isolated from immunoprecipitates and then RT-PCR was performed.
ChIP Assay
Cross-linked cells were washed with PBS, resuspended in lysis buffer, and sonicated. Chromatin extracts are precleared with protein-A/G-Sepharose beads and then immunoprecipitated with antibodies on protein-A/G-Sepharose beads. After washing, elution, and de-cross-linking, the ChIP DNA was detected by PCR.
Super-EMSA (Gel-Shift)
Cells were washed and scraped in ice-cold PBS to prepare nuclei for electrophoretic gel mobility shift assay with the use of the gel shift assay system modified according to the manufacturer’s instructions (Promega).
Dual Luciferase Reporter Assay
Cells were transiently transfected with luciferase construct with the use of Lipofectamine 2000 (Invitrogen). After incubation for 36 hr, the cells were harvested with passive lysis buffer, and luciferase activities of cell extracts were measured with the use of the Dual Luciferase Assay system (Promega) according to manufacturer’s instructions.
Cell Proliferation CCK8 Assay
The cell proliferation is measured using CCK8 Assay kit according to the manufacturer’s instructions (Boshide, Wuhan, China).
Colony Formation Efficiency Assay
Cells were plated on dishes, and the DMEM containing 10% FBS was added into each dish. Cell colonies on the dishes were stained with 0.5% Crystal Violet for more than 1 hr, and the colonies were counted.
Xenograft Transplantation In Vivo
Four-week-old athymic Balb/C mice were purchased from Shi Laike Company (Shanghai, China) and maintained in the Tongji animal facilities approved by the China Association for Accreditation of Laboratory Animal Care. The athymic Balb/C mice were injected in the armpit area subcutaneously with liver cancer cells. The mice were observed four weeks and then sacrificed to recover the tumors.
Statistical Analysis
The significant differences between mean values were obtained from at least three independent experiments. Each value was presented as mean ± SEM. Student’s t test was used for comparisons, with p < 0.05 or p < 0.01 considered significant.
Author Contributions
D.L. conceived the study and participated in the study design, performance, coordination, and manuscript writing. Z.L., Y.L., Q.M., C.W., X.L., Y.Y., X.X., Q.Z., J.X., X.G., T.L., H.P., W.X., J.L., and S.J. performed the research. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (NCSF; 81572773 and 81773158) and the Key Specialty Construction Project of Pudong Health and Family Planning Commission of Shanghai (PWZz2013-05).
Footnotes
Supplemental Information includes Supplemental Results and four figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.04.001.
Supplemental Information
References
- 1.Chen X., Hao B., Han G., Liu Y., Dai D., Li Y., Wu X., Zhou X., Yue Z., Wang L. miR-372 regulates glioma cell proliferation and invasion by directly targeting PHLPP2. J. Cell. Biochem. 2015;116:225–232. doi: 10.1002/jcb.24949. [DOI] [PubMed] [Google Scholar]
- 2.Tu H.F., Chang K.W., Cheng H.W., Liu C.J. Upregulation of miR-372 and -373 associates with lymph node metastasis and poor prognosis of oral carcinomas. Laryngoscope. 2015;125:E365–E370. doi: 10.1002/lary.25464. [DOI] [PubMed] [Google Scholar]
- 3.Zhou C., Li X., Zhang X., Liu X., Tan Z., Yang C., Zhang J. microRNA-372 maintains oncogene characteristics by targeting TNFAIP1 and affects NFκB signaling in human gastric carcinoma cells. Int. J. Oncol. 2013;42:635–642. doi: 10.3892/ijo.2012.1737. [DOI] [PubMed] [Google Scholar]
- 4.Feng L., Ma Y., Sun J., Shen Q., Liu L., Lu H., Wang F., Yue Y., Li J., Zhang S. YY1-MIR372-SQSTM1 regulatory axis in autophagy. Autophagy. 2014;10:1442–1453. doi: 10.4161/auto.29486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X., Zhang X., Liu X., Tan Z., Yang C., Ding X., Hu X., Zhou J., Xiang S., Zhou C., Zhang J. Caudatin induces cell apoptosis in gastric cancer cells through modulation of Wnt/β-catenin signaling. Oncol. Rep. 2013;30:677–684. doi: 10.3892/or.2013.2495. [DOI] [PubMed] [Google Scholar]
- 6.Tran N.D., Kissner M., Subramanyam D., Parchem R.J., Laird D.J., Blelloch R.H. A miR-372/let-7 axis regulates human germ versus somatic cell fates. Stem Cells. 2016;34:1985–1991. doi: 10.1002/stem.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B.L., Sun K.X., Zong Z.H., Chen S., Zhao Y. MicroRNA-372 inhibits endometrial carcinoma development by targeting the expression of the Ras homolog gene family member C (RhoC) Oncotarget. 2016;7:6649–6664. doi: 10.18632/oncotarget.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh L.Y., Liu C.J., Wong Y.K., Chang C., Lin S.C., Chang K.W. miR-372 inhibits p62 in head and neck squamous cell carcinoma in vitro and in vivo. Oncotarget. 2015;6:6062–6075. doi: 10.18632/oncotarget.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao H., Feng Y., Chen L. Repression of microRNA-372 by arsenic sulphide inhibits prostate cancer cell proliferation and migration through regulation of large tumour suppressor kinase 2. Basic Clin. Pharmacol. Toxicol. 2017;120:256–263. doi: 10.1111/bcpt.12687. [DOI] [PubMed] [Google Scholar]
- 10.Tan J.K., Tan E.L., Gan S.Y. Elucidating the roles of miR-372 in cell proliferation and apoptosis of nasopharyngeal carcinoma TW01 cells. Exp. Oncol. 2014;36:170–173. [PubMed] [Google Scholar]
- 11.Guan X., Zong Z.H., Chen S., Sang X.B., Wu D.D., Wang L.L., Liu Y., Zhao Y. The role of miR-372 in ovarian carcinoma cell proliferation. Gene. 2017;624:14–20. doi: 10.1016/j.gene.2017.04.043. [DOI] [PubMed] [Google Scholar]
- 12.Chen H., Zhang Z., Lu Y., Song K., Liu X., Xia F., Sun W. Downregulation of ULK1 by microRNA-372 inhibits the survival of human pancreatic adenocarcinoma cells. Cancer Sci. 2017;108:1811–1819. doi: 10.1111/cas.13315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou W., Yuan T., Gao Y., Yin P., Liu W., Pan C., Liu Y., Yu X. IL-1β-induces NF-κB and upregulates microRNA-372 to inhibit spinal cord injury recovery. J. Neurophysiol. 2017;117:2282–2291. doi: 10.1152/jn.00936.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong X., Qian X., Duan L., Liu H., Zhu Y., Qi J. MicroRNA-372 suppresses migration and invasion by targeting p65 in human prostate cancer cells. DNA Cell Biol. 2016;35:828–835. doi: 10.1089/dna.2015.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J., Jin L., Jiang L., Gao L., Zhou J., Hu Y., Li W., Zhi Q., Zhu X. Serum miR-372 is a diagnostic and prognostic biomarker in patients with early colorectal cancer. Anticancer. Agents Med. Chem. 2016;16:424–431. doi: 10.2174/1871520615666150716110406. [DOI] [PubMed] [Google Scholar]
- 16.Huang X., Huang M., Kong L., Li Y. miR-372 suppresses tumour proliferation and invasion by targeting IGF2BP1 in renal cell carcinoma. Cell Prolif. 2015;48:593–599. doi: 10.1111/cpr.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu G., Liu H., He H., Wang Y., Lu X., Yu Y., Xia S., Meng X., Liu Y. miR-372 down-regulates the oncogene ATAD2 to influence hepatocellular carcinoma proliferation and metastasis. BMC Cancer. 2014;14:107. doi: 10.1186/1471-2407-14-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu G., Wang Y., Lu X., He H., Liu H., Meng X., Xia S., Zheng K., Liu B. Low mir-372 expression correlates with poor prognosis and tumor metastasis in hepatocellular carcinoma. BMC Cancer. 2015;15:182. doi: 10.1186/s12885-015-1214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi J.H., Cui N.P., Wang S., Zhao M.Z., Wang B., Wang Y.N., Chen B.P. Overexpression of YB1 C-terminal domain inhibits proliferation, angiogenesis and tumorigenicity in a SK-BR-3 breast cancer xenograft mouse model. FEBS Open Bio. 2016;6:33–42. doi: 10.1002/2211-5463.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotake Y., Arikawa N., Tahara K., Maru H., Naemura M. Y-box binding protein 1 is involved in regulating the G2/M phase of the cell cycle. Anticancer Res. 2017;37:1603–1608. doi: 10.21873/anticanres.11490. [DOI] [PubMed] [Google Scholar]
- 21.Li C.X., Yu B., Shi L., Geng W., Lin Q.B., Ling C.C., Yang M., Ng K.T., Huang J.D., Man K. ‘Obligate’ anaerobic Salmonella strain YB1 suppresses liver tumor growth and metastasis in nude mice. Oncol. Lett. 2017;13:177–183. doi: 10.3892/ol.2016.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia J., Zheng Y., Wang W., Shao Y., Li Z., Wang Q., Wang Y., Yan H. Antimicrobial peptide LL-37 promotes YB-1 expression, and the viability, migration and invasion of malignant melanoma cells. Mol. Med. Rep. 2017;15:240–248. doi: 10.3892/mmr.2016.5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mo D., Fang H., Niu K., Liu J., Wu M., Li S., Zhu T., Aleskandarany M.A., Arora A., Lobo D.N. Human helicase RECQL4 drives cisplatin resistance in gastric cancer by activating an AKT-YB1-MDR1 signaling pathway. Cancer Res. 2016;76:3057–3066. doi: 10.1158/0008-5472.CAN-15-2361. [DOI] [PubMed] [Google Scholar]
- 24.Dey A., Robitaille M., Remke M., Maier C., Malhotra A., Gregorieff A., Wrana J.L., Taylor M.D., Angers S., Kenney A.M. YB-1 is elevated in medulloblastoma and drives proliferation in Sonic hedgehog-dependent cerebellar granule neuron progenitor cells and medulloblastoma cells. Oncogene. 2016;35:4256–4268. doi: 10.1038/onc.2015.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu T.T., Arango-Argoty G., Li Z., Lin Y., Kim S.W., Dueck A., Ozsolak F., Monaghan A.P., Meister G., DeFranco D.B., John B. Noncoding RNAs that associate with YB-1 alter proliferation in prostate cancer cells. RNA. 2015;21:1159–1172. doi: 10.1261/rna.045559.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiota M., Bishop J.L., Takeuchi A., Nip K.M., Cordonnier T., Beraldi E., Kuruma H., Gleave M.E., Zoubeidi A. Inhibition of the HER2-YB1-AR axis with lapatinib synergistically enhances enzalutamide anti-tumor efficacy in castration resistant prostate cancer. Oncotarget. 2015;6:9086–9098. doi: 10.18632/oncotarget.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X., Ding Z., Mo J., Sang B., Shi Q., Hu J., Xie S., Zhan W., Lu D., Yang M. GOLPH3 promotes glioblastoma cell migration and invasion via the mTOR-YB1 pathway in vitro. Mol. Carcinog. 2015;54:1252–1263. doi: 10.1002/mc.22197. [DOI] [PubMed] [Google Scholar]
- 28.Song Y.J., Lee H. YB1/p32, a nuclear Y-box binding protein 1, is a novel regulator of myoblast differentiation that interacts with Msx1 homeoprotein. Exp. Cell Res. 2010;316:517–529. doi: 10.1016/j.yexcr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Xiong H., Ni Z., He J., Jiang S., Li X., He J., Gong W., Zheng L., Chen S., Li B. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528–3540. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 30.Li D., Liu X., Zhou J., Hu J., Zhang D., Liu J., Qiao Y., Zhan Q. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology. 2017;65:1612–1627. doi: 10.1002/hep.29010. [DOI] [PubMed] [Google Scholar]
- 31.Wang W.T., Ye H., Wei P.P., Han B.W., He B., Chen Z.H., Chen Y.Q. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J. Hematol. Oncol. 2016;9:117. doi: 10.1186/s13045-016-0348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y., Xu S., Jin W., Yi Q., Wei W. Effect of the PTEN gene on adhesion, invasion and metastasis of osteosarcoma cells. Oncol. Rep. 2014;32:1741–1747. doi: 10.3892/or.2014.3362. [DOI] [PubMed] [Google Scholar]
- 33.Li X.Y., Wang S.S., Han Z., Han F., Chang Y.P., Yang Y., Xue M., Sun B., Chen L.M. Triptolide restores autophagy to alleviate diabetic renal fibrosis through the miR-141-3p/PTEN/Akt/mTOR pathway. Mol. Ther. Nucleic Acids. 2017;9:48–56. doi: 10.1016/j.omtn.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z., Li J., Bi P., Lu Y., Burcham G., Elzey B.D., Ratliff T., Konieczny S.F., Ahmad N., Kuang S., Liu X. Plk1 phosphorylation of PTEN causes a tumor-promoting metabolic state. Mol. Cell. Biol. 2014;34:3642–3661. doi: 10.1128/MCB.00814-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong S., Cheng J.C., Klausen C., Zhao J., Leung P.C. TGF-β1 stimulates migration of type II endometrial cancer cells by down-regulating PTEN via activation of SMAD and ERK1/2 signaling pathways. Oncotarget. 2016;7:61262–61272. doi: 10.18632/oncotarget.11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao L.Y., Tong D.D., Xue M., Ma H.L., Liu S.Y., Yang J., Liu Y.X., Guo B., Ni L., Liu L.Y. MeCP2, a target of miR-638, facilitates gastric cancer cell proliferation through activation of the MEK1/2-ERK1/2 signaling pathway by upregulating GIT1. Oncogenesis. 2017;6:e368. doi: 10.1038/oncsis.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuijers J., Mokry M., Hatzis P., Cuppen E., Clevers H. Wnt-induced transcriptional activation is exclusively mediated by TCF/LEF. EMBO J. 2014;33:146–156. doi: 10.1002/embj.201385358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh S., Kim H., Nam K., Shin I. Silencing of Glut1 induces chemoresistance via modulation of Akt/GSK-3β/β-catenin/survivin signaling pathway in breast cancer cells. Arch. Biochem. Biophys. 2017;636:110–122. doi: 10.1016/j.abb.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Dong L., Li Z., Xue L., Li G., Zhang C., Cai Z., Li H., Guo R. DIAPH3 promoted the growth, migration and metastasis of hepatocellular carcinoma cells by activating beta-catenin/TCF signaling. Mol. Cell. Biochem. 2018;438:183–190. doi: 10.1007/s11010-017-3125-7. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J.H., Jiao L.Y., Li T.J., Zhu Y.Y., Zhou J.W., Tian J. GSK-3β suppresses HCC cell dissociation in vitro by upregulating epithelial junction proteins and inhibiting Wnt/β-catenin signaling pathway. J. Cancer. 2017;8:1598–1608. doi: 10.7150/jca.18744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain S., Ghanghas P., Rana C., Sanyal S.N. Role of GSK-3β in regulation of canonical Wnt/β-catenin signaling and PI3-K/Akt oncogenic pathway in colon cancer. Cancer Invest. 2017;35:473–483. doi: 10.1080/07357907.2017.1337783. [DOI] [PubMed] [Google Scholar]
- 42.Chandra V., Fatima I., Manohar M., Popli P., Sirohi V.K., Hussain M.K., Hajela K., Sankhwar P., Dwivedi A. Inhibitory effect of 2-(piperidinoethoxyphenyl)-3-(4-hydroxyphenyl)-2H-benzo(b)pyran (K-1) on human primary endometrial hyperplasial cells mediated via combined suppression of Wnt/β-catenin signaling and PI3K/Akt survival pathway. Cell Death Dis. 2014;5:e1380. doi: 10.1038/cddis.2014.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang J., Cao R., Wang X., Zhang Y., Wang P., Gao H., Li C., Yang F., Zeng R., Wei P. Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2. Cell Res. 2017;27:329–351. doi: 10.1038/cr.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C., Jiang J., Ji J., Cai Q., Chen X., Yu Y., Zhu Z., Zhang J. PKM2 promotes cell migration and inhibits autophagy by mediating PI3K/AKT activation and contributes to the malignant development of gastric cancer. Sci. Rep. 2017;7:2886. doi: 10.1038/s41598-017-03031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tee S.S., Park J.M., Hurd R.E., Brimacombe K.R., Boxer M.B., Massoud T.F., Rutt B.K., Spielman D.M. PKM2 activation sensitizes cancer cells to growth inhibition by 2-deoxy-D-glucose. Oncotarget. 2017;8:90959–90968. doi: 10.18632/oncotarget.19630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Méndez-Lucas A., Li X., Hu J., Che L., Song X., Jia J., Wang J., Xie C., Driscoll P.C., Tschaharganeh D.F. Glucose catabolism in liver tumors induced by c-MYC can be sustained by various PKM1/PKM2 ratios and pyruvate kinase activities. Cancer Res. 2017;77:4355–4364. doi: 10.1158/0008-5472.CAN-17-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei L., Dai Y., Zhou Y., He Z., Yao J., Zhao L., Guo Q., Yang L. Oroxylin A activates PKM1/HNF4 alpha to induce hepatoma differentiation and block cancer progression. Cell Death Dis. 2017;8:e2944. doi: 10.1038/cddis.2017.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morfouace M., Lalier L., Oliver L., Cheray M., Pecqueur C., Cartron P.F., Vallette F.M. Control of glioma cell death and differentiation by PKM2-Oct4 interaction. Cell Death Dis. 2014;5:e1036. doi: 10.1038/cddis.2013.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gates L.A., Shi J., Rohira A.D., Feng Q., Zhu B., Bedford M.T., Sagum C.A., Jung S.Y., Qin J., Tsai M.J. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J. Biol. Chem. 2017;292:14456–14472. doi: 10.1074/jbc.M117.802074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lutz L., Fitzner I.C., Ahrens T., Geißler A.L., Makowiec F., Hopt U.T., Bogatyreva L., Hauschke D., Werner M., Lassmann S. Histone modifiers and marks define heterogeneous groups of colorectal carcinomas and affect responses to HDAC inhibitors in vitro. Am. J. Cancer Res. 2016;6:664–676. [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Q., Tong C., Luo M., Xue X., Mei Q., Ma L., Yu X., Mao W., Kong L., Yu X., Li S. Regulation of SESAME-mediated H3T11 phosphorylation by glycolytic enzymes and metabolites. PLoS ONE. 2017;12:e0175576. doi: 10.1371/journal.pone.0175576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z. ErbB receptors and cancer. Methods Mol. Biol. 2017;1652:3–35. doi: 10.1007/978-1-4939-7219-7_1. [DOI] [PubMed] [Google Scholar]
- 53.Simond A.M., Rao T., Zuo D., Zhao J.J., Muller W.J. ErbB2-positive mammary tumors can escape PI3K-p110α loss through downregulation of the Pten tumor suppressor. Oncogene. 2017;36:6059–6066. doi: 10.1038/onc.2017.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hasegawa T., Adachi R., Iwakata H., Takeno T., Sato K., Sakamaki T. ErbB2 signaling epigenetically suppresses microRNA-205 transcription via Ras/Raf/MEK/ERK pathway in breast cancer. FEBS Open Bio. 2017;7:1154–1165. doi: 10.1002/2211-5463.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuda H., Campion C.G., Fujiwara K., Ikeda J., Cossette S., Verissimo T., Ogasawara M., Gaboury L., Saito K., Yamaguchi K. HCaRG/COMMD5 inhibits ErbB receptor-driven renal cell carcinoma. Oncotarget. 2017;8:69559–69576. doi: 10.18632/oncotarget.18012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qi L., Zhang B., Zhang S., Ci X., Wu Q., Ma G., Luo A., Fu L., King J.L., Nahta R., Dong J.T. ERRF sensitizes ERBB2-positive breast cancer cells to lapatinib treatment likely by attenuating MCL1 and ERBB2 expression. Oncotarget. 2017;8:36054–36066. doi: 10.18632/oncotarget.16425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Z., Oyesanya R.A., Campbell D.J., Almenara J.A., Dewitt J.L., Sirica A.E. Preclinical assessment of simultaneous targeting of epidermal growth factor receptor (ErbB1) and ErbB2 as a strategy for cholangiocarcinoma therapy. Hepatology. 2010;52:975–986. doi: 10.1002/hep.23773. [DOI] [PubMed] [Google Scholar]
- 58.Liu J., Ahiekpor A., Li L., Li X., Arbuthnot P., Kew M., Feitelson M.A. Increased expression of ErbB-2 in liver is associated with hepatitis B x antigen and shorter survival in patients with liver cancer. Int. J. Cancer. 2009;125:1894–1901. doi: 10.1002/ijc.24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C., Ge Q., Chen Z., Hu J., Li F., Ye Z. Promoter-associated endogenous and exogenous small RNAs suppress human bladder cancer cell metastasis by activating p21 (CIP1/WAF1) expression. Tumour Biol. 2016;37:6589–6598. doi: 10.1007/s13277-015-4571-z. [DOI] [PubMed] [Google Scholar]
- 60.Wang C., Tang K., Li Z., Chen Z., Xu H., Ye Z. Targeted p21(WAF1/CIP1) activation by miR-1236 inhibits cell proliferation and correlates with favorable survival in renal cell carcinoma. Urol. Oncol. 2016;34:59.e23–59.e34. doi: 10.1016/j.urolonc.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 61.Tang K., Wang C., Chen Z., Xu H., Ye Z. Clinicopathologic and prognostic significance of p21 (Cip1/Waf1) expression in bladder cancer. Int. J. Clin. Exp. Pathol. 2015;8:4999–5007. [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao J., Lammers P., Torrance C.J., Bader A.G. TP53-independent function of miR-34a via HDAC1 and p21(CIP1/WAF1.) Mol. Ther. 2013;21:1678–1686. doi: 10.1038/mt.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Y.F., Wang C.R., Wu Y.M., Ma S.L., Ji Y., Lu Y.J. P21 (waf1/cip1) is required for non-small cell lung cancer sensitive to Gefitinib treatment. Biomed. Pharmacother. 2011;65:151–156. doi: 10.1016/j.biopha.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 64.Peng C., Zeng W., Su J., Kuang Y., He Y., Zhao S., Zhang J., Ma W., Bode A.M., Dong Z., Chen X. Cyclin-dependent kinase 2 (CDK2) is a key mediator for EGF-induced cell transformation mediated through the ELK4/c-Fos signaling pathway. Oncogene. 2016;35:1170–1179. doi: 10.1038/onc.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai L., Liu Y., Liu J., Wen X., Xu Z., Wang Z., Sun H., Tang S., Maguire A.R., Quan J. A novel cyclinE/cyclinA-CDK inhibitor targets p27(Kip1) degradation, cell cycle progression and cell survival: implications in cancer therapy. Cancer Lett. 2013;333:103–112. doi: 10.1016/j.canlet.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 66.Liu B., Wen J.K., Li B.H., Fang X.M., Wang J.J., Zhang Y.P., Shi C.J., Zhang D.Q., Han M. Celecoxib and acetylbritannilactone interact synergistically to suppress breast cancer cell growth via COX-2-dependent and -independent mechanisms. Cell Death Dis. 2011;2:e185. doi: 10.1038/cddis.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Z., Li G., Bian E., Ma C.C., Wan J., Zhao B. TGF-β-mediated repression of MST1 by DNMT1 promotes glioma malignancy. Biomed. Pharmacother. 2017;94:774–780. doi: 10.1016/j.biopha.2017.07.081. [DOI] [PubMed] [Google Scholar]
- 68.Hu Y., Ma Z., He Y., Liu W., Su Y., Tang Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem. Biophys. Res. Commun. 2017;491:926–931. doi: 10.1016/j.bbrc.2017.07.137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.