Abstract
RNA modifications are additions of chemical groups to nucleotides or their local structural changes. Knowledge about the occurrence sites of these modifications is essential for in-depth understanding of the biological functions and mechanisms and for treating some genomic diseases as well. With the avalanche of RNA sequences generated in the post-genomic age, many computational methods have been proposed for identifying various types of RNA modifications one by one. However, so far no method whatsoever has been developed for simultaneously identifying several different types of RNA modifications. To address such a challenge, we developed a predictor called “iRNA-3typeA,” by which we can simultaneously identify the occurrence sites of the following three most frequently observed modifications in RNA: (1) N1-methyladenosine (m1A), (2) N6-methyladenosine (m6A), and (3) adenosine to inosine (A-to-I). It has been shown via rigorous cross-validations for the RNA sequences from Homo sapiens and Mus musculus transcriptomes that the success rates achieved by the powerful new predictor are quite high. For the convenience of broad experimental scientists, a user-friendly web server for iRNA-3typeA has been established at http://lin-group.cn/server/iRNA-3typeA/. It is anticipated that iRNA-3typeA may become a useful high throughput tool for genome analysis.
Keywords: RNA modification, N1-methyladenosine, N6-methyladenosine, adenosine to inosine editing, five-step rules, web server
Introduction
RNA modification means the addition of chemical groups to its constitutional nucleotides or structural changes therein.1 So far, more than 100 types of RNA modifications have been observed in cellular RNAs of all living organisms.2 Because they are involved in a series of crucial biological activities,3 such as mRNA splicing, mRNA nuclear processing, mRNA export, and mRNA decay,3, 4, 5, 6 particularly linked with human diseases, RNA modifications have drawn great attention in the scientific community.
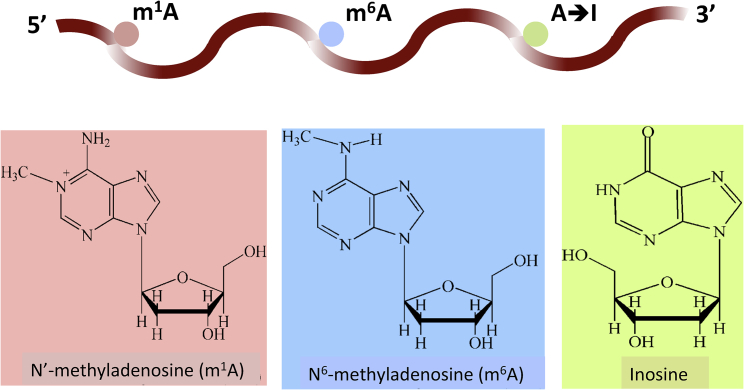
With the development of high-throughput experimental techniques,7, 8, 9 lots of RNA modification data have been acquired; they are very helpful for revealing the novel functions of RNA modifications. As indicated in a recent review,10 however, most of these methods are unable to discriminate among the different RNA modifications that may simultaneously occur in the same RNA molecule. For example, the adenosine usually undergoes N1-methyladenosine (m1A), N6-methyladenosine (m6A), and adenosine to inosine (A-to-I or ) modifications7 (Figure 1). Unfortunately, using the aforementioned techniques, one could not detect whether different types of RNA modifications might take place at the same time, let alone analyze their combinational biological functions.11.
Figure 1.
The Three Common Types of Modifications in RNA
(1) N1-methyladenosine (m1A), (2) N6-methyladenosine (m6A), and (3) adenosine to inosine (A-to-I).
Therefore, it is urgently needed to develop computational methods to address this problem. As excellent complements to experimental techniques, computational methods have been developed to identify RNA modifications12, 13, 14, 15, 16, 17, 18 via machine learning to train computational models based on the large data yielded from the high-throughput experiments. However, rarely are they able to simultaneously identify multiple RNA modifications.
The present study was devoted to developing a bioinformatics tool that can identify the RNA modification types for m1A, m6A, and that may simultaneously occur on adenosine in both Homo sapiens and Mus musculus transcriptomes.
As shown in a series of recent publications,19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 in developing a bioinformatics tool, complying with the five-step rules yields the following advantages:32 (1) clearer in logic deduction, (2) better illumination in stimulating other relevant tools, and (3) more usefulness in practical application.
In view of this, we elaborate the following procedures required in the five-step rules: (1) benchmark dataset, (2) sample formulation, (3) operative machine, (4) cross-validation, and (5) web server, and they are embedded into the rubrics according to the journal’s format.
Results and Discussion
Performance Report
Listed in Table 1 are the jackknife test results obtained by the proposed predictor on the benchmark datasets (Supplemental Information S1 and Supplemental Information S2 available at http://lin-group.cn/server/iRNA3typeA/data.htm) for H. sapiens and M. musculus, respectively. As we can see from the table, the rates for both overall accuracy (Acc) and stability (MCC) are quite high for all the three different types of modifications investigated, indicating that the predictor is not only high in overall success rate but also quite stable. Therefore, the potential is quite high for iRNA-type3A to become a high-throughput tool in both basic research and drug development.
Table 1.
The Success Rates Achieved by iRNA-3typeA via Jackknife Tests on the Benchmark Datasets for H. sapiens and M. musculus, Respectively
| Species | Type of Modification | Sn (%) | Sp (%) | Acc (%) | MCC |
|---|---|---|---|---|---|
| H. sapiens | m1Aa | 98.38 | 99.89 | 99.13 | 0.98 |
| m6Ab | 81.68 | 99.11 | 90.38 | 0.82 | |
| c | 86.18 | 95.23 | 90.71 | 0.82 | |
| M. musculus | m1Ad | 97.46 | 100.00 | 98.73 | 0.97 |
| m6Ae | 77.79 | 100.00 | 88.39 | 0.80 | |
| f | 96.75 | 100.00 | 98.38 | 0.96 |
The parameters used for SVM are and = 0.0078125.
The parameters used for SVM are and = 3.05158e-5.
The parameters used for SVM are and = 0.0078125.
The parameters used for SVM are and = 0.0078125.
The parameters used for SVM are and = 0.00012207.
The parameters used for SVM are and = 0.000488281.
It is instructive to point out that, although the current predictor is limited in identifying m1A, m6A, and sites for the RNA sequences from H. sapiens and M. musculus, with more experimental data available for other types of modifications and other species in future, we can easily to extend our model to cover more different types of modifications and more different species. Therefore, the current predictor is just a good start; it will be subjected to updates with the aim to continuously enhance its power and coverage scope.
Comparison with Other Classifiers
The proposed predictor iRNA-3typeA is the first predictor ever constructed for identifying the three types of RNA modifications (m1A; m6A; ) simultaneously. It is not possible to show its power via a conventional comparison since there is no other predictor whatsoever that can do the same. Nevertheless, below we can carry out a special comparison to further demonstrate its superiority.
As mentioned above, the operative machine used for iRNA-3typeA is a support vector machine (SVM) classifier. What would happen if we use other classifiers instead? Listed in Table 2 are the results when the SVM classifier was substituted with the other classifiers, respectively.
Table 2.
The Comparative Results of the Proposed Predictor When Its Operating Algorithm32 Was Replaced from SVM to Other Classifiers
| Classifier | Species | Modification Type | Sn (%) | Sp (%) | Acc (%) | MCC |
|---|---|---|---|---|---|---|
| BayesNeta | H. sapiens | m1A | 98.81 | 98.85 | 98.83 | 0.98 |
| m6A | 82.04 | 100.00 | 91.02 | 0.83 | ||
| 88.50 | 89.57 | 89.03 | 0.78 | |||
| M. musculus | m1A | 97.18 | 98.78 | 97.98 | 0.96 | |
| m6A | 77.79 | 100.00 | 88.90 | 0.80 | ||
| 96.51 | 99.88 | 98.20 | 0.96 | |||
| Naive Bayesa | H. sapiens | m1A | 98.16 | 98.30 | 98.23 | 0.96 |
| m6A | 82.04 | 99.73 | 90.88 | 0.83 | ||
| 89.40 | 87.04 | 88.22 | 0.76 | |||
| M. musculus | m1A | 96.43 | 97.75 | 97.09 | 0.94 | |
| m6A | 77.79 | 98.62 | 88.22 | 0.78 | ||
| 95.91 | 97.95 | 96.93 | 0.94 | |||
| J48 Treea | H. sapiens | m1A | 98.77 | 99.40 | 99.09 | 0.98 |
| m6A | 82.48 | 84.35 | 83.41 | 0.67 | ||
| 88.18 | 89.04 | 88.60 | 0.77 | |||
| M. musculus | m1A | 96.71 | 98.68 | 97.70 | 0.95 | |
| m6A | 83.03 | 82.21 | 82.62 | 0.65 | ||
| 96.27 | 99.04 | 97.65 | 0.95 | |||
| SVMb | H. sapiens | m1A | 98.46 | 99.89 | 99.18 | 0.98 |
| m6A | 80.44 | 100.00 | 90.23 | 0.82 | ||
| 86.73 | 95.40 | 91.07 | 0.82 | |||
| M. musculus | m1A | 97.46 | 100.00 | 98.73 | 0.97 | |
| m6A | 77.79 | 100.00 | 88.90 | 0.80 | ||
| 97.35 | 100.00 | 98.67 | 0.97 |
All the rates below are obtained by the 10-fold cross-validations on the same benchmark datasets (Supplemental Information S1 and Supplemental Information S2 available at http://lin-group.cn/server/iRNA3typeA/data.htm).
Taken from the WEKA package.91
Proposed in this paper.
From the table, we can see the following: (1) the SVM classifier is better than J48 Tree in all the metrics rates. (2) Although the SVM classifier is a little bit lower than the BayesNet classifier and Naive Bayes classifier in identifying the m6A sites for H. sapiens, its accuracies in identifying all the other types of modifications for both H. sapiens and M. musculus are significantly higher than those of BayesNet and Naive Bayes. All these results have further indicated that the SVM classifier is indeed a correct choice for the iRNA-3typeA predictor.
Web Server and User Guide
The last step of the five-step rules32 is about the web server. It is indeed important because user-friendly and publicly accessible web servers represent the future direction for developing practically more useful predictors.33 Actually, it has been demonstrated by a series of recent publications (see, e.g., Cheng et al.,25, 34, 35, 36 Liu et al.,28 Lin et al.,37 Jia et al.,38, 39 and Cheng and Xiao40) that a new prediction method with its web server available would significantly enhance its impacts.41, 42 In view of this, the web server for iRNA-3typeA has been established. Furthermore, to maximize the convenience of broad experimental scientists, a step-by-step guide is given below:
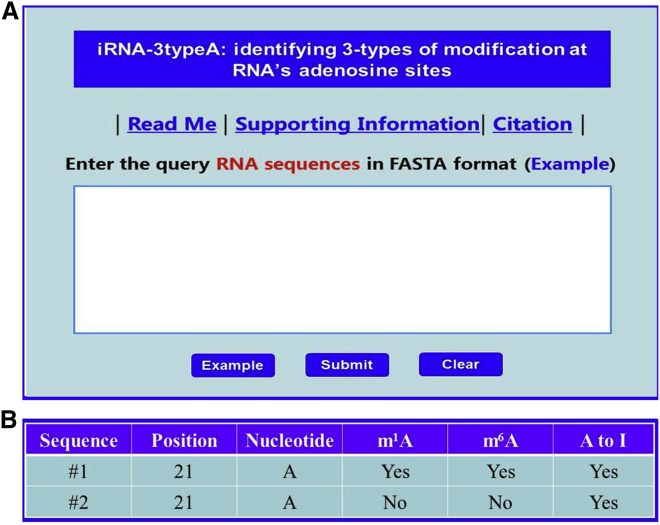
Step 1. Open the iRNA-3typeA web server at http://lin-group.cn/server/iRNA-3typeA; you will see the top page of the web server as shown in Figure 2A.
Step 2. Either type or copy/paste the query RNA sequences (in FASTA format) into the input box. Example sequences can be found by clicking on the Example button.
Step 3. Click the open circle (H. sapiens and M. musculus) to choose the species concerned, followed by clicking the Submit button. For example, if using the query RNA sequences in the Example window as the input and choosing H. sapiens, after submission you will see the predicted results summarized in a table (Figure 2B), clearly indicating (1) the adenosine at position 21of sequence #1 has the potential to be of the site for m1A or A-to-I editing modification. (2) The adenosine at position 21 of sequence #2 has the potential to be of m6A modification only. All these predicted results are fully consistent with experimental observations.
Figure 2.
The Semi-screenshot for the Top Page of the iRNA-3typeA Web Server and the Prediction Result of the Two Example Query Sequences
The Semi-screenshot for the top page of the iRNA-3typeA Web Server (top panel) and the Prediction Result of the two example query sequences (bottom panel).
Materials and Methods
Benchmark Datasets
The benchmark datasets for m1A, m6A, and A-to-I editing sites in H. sapiens and M. musculus genomes were derived from the previous works.12, 14, 43 Listed in Table 3 are the numbers of positive and negative samples for each of the benchmark datasets. It has been found by similar approaches12, 14 that the optimal length of the sequence samples in the benchmark datasets are 41nt, with the modified sites (m1A, m6A, or editing site) at the center. For readers’ convenience, the benchmark dataset thus obtained for H. sapiens is given in Supplemental Information S1, while that for M. musculus given in Supplemental Information S2; both can be downloaded from the link at http://lin-group.cn/server/iRNA3typeA/data.htm.
Table 3.
A Breakdown of the Benchmark Dataset
| Species | Attribute | Number of Samples |
||
|---|---|---|---|---|
| m1A | m6A | |||
| H. sapiens | positive | 6,366 | 1,130 | 3,000 |
| negative | 6,366 | 1,130 | 3,000 | |
| M. musculus | positive | 1,064 | 725 | 831 |
| negative | 1,064 | 725 | 831 | |
Sample Formulation
An RNA sample with 41 nt is usually sequentially formulated by
| (Equation 1) |
where
| (Equation 2) |
denotes the nucleotide at the i-th sequence position, and is the a symbol in the set theory meaning “member of.”
To enable the existing machine-learning algorithms handle the RNA sample,41 the first thing we need to do is to convert its sequential formulation into a vector. But a vector in a discrete framework might totally miss all the sequence-order information or pattern feature. To deal with this problem, the PseAAC (pseudo amino acid composition) was introduced.44 Ever since the concept of PseAAC was proposed, it has been swiftly penetrated into many biomedicine and drug development areas45, 46 and nearly all the areas of computational proteomics (see, e.g.,Esmaeili et al.,47 Mohabatkar et al.,48 Nanni et al.,49 Pacharawongsakda and Theeramunkong,50 Mondal and Pai,51 Ahman et al.,52 Kabir and Hayat,53 Yu et al.,54 Zhang and Duan,55 Muthu Krishnan,56 and a long list of references cited in two review papers42, 57). Encouraged by the successes of using PseAAC to deal with protein/peptide sequences, this idea has been extended to deal with DNA/RNA sequences21, 28, 37, 58, 59, 60 in computational genomics via PseKNC (pseudo K-tuple nucleotide composition).61, 62 According to Chen et al.63, the general form of PseKNC can be formulated as
| (Equation 3) |
where T is the transposing operator, the subscript is an integer, and its value and the components will depend on how to extract the desired features and properties from the RNA sequence (cf. Equation 1). In this study, their definitions are described below.
The four bases (A, C, G, and U) of RNA have different chemical properties and structures.64, 65 Therefore, based on their different chemical properties and structures,64, 65 A, C, G, and U can be represented by (1, 1, 1), (0, 0, 1), (1, 0, 0), and (0, 1, 0), respectively.20, 27 For instance, the RNA sequence with six nucleotides “GUGCAG” can be expressed by the vector of components; i.e., [1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0, 1, 1, 1, 1, 1, 0, 0].
Moreover, to incorporate into Equation 3 the sequence-coupled information66 for the nucleotides around the modification sites, we adopt the lingering density as defined below
| (Equation 4) |
where is the density of the nucleotide at the site of a RNA sequence, the length of the sliding substring concerned; denotes each of the site locations counted in the substring, and
| (Equation 5) |
For example, the RNA sequence “GUGCAG” can be represented by the vector [1, 0.5, 0.66, 0.25, 0.2, 0.5].
Thus, by using both nucleotide chemical properties and the lingering density (cf. Equation 4), each nucleotide can be defined by four variables. Accordingly, the RNA sequence of Equation 1 can be defined by a vector with components; namely for Equation 3 now.
Operative Machine
In this study, the SVM was chosen as the operative machine. The SVM has been widely used in computational genomics and proteomics (see, e.g., Ehsan et al.,26 Feng et al.,20, 27, 67, 68, 69 Chen et al.,70, 71, 72 Lin et al.,73 Lai et al.,74 Zhao et al.,75 and Yang et al.76). The implementation of the SVM was conducted by using the LibSVM package 3.18 available at https://www.csie.ntu.edu.tw/∼cjlin/libsvm/. The radial basis kernel function (RBF) was used to obtain the classification hyperplane, and the grid search method was applied to optimize the regularization parameter C and kernel parameter γ.
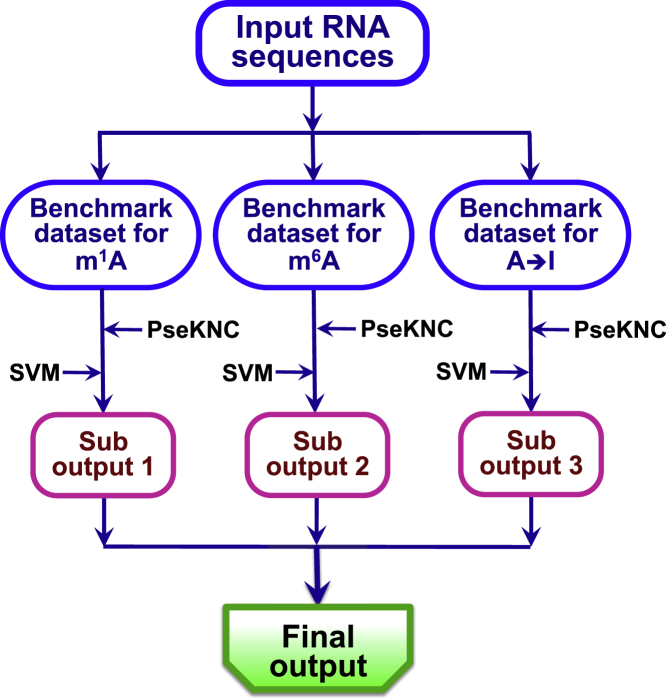
The predictor obtained via the above procedures is called “iRNA-3typeA,” where “i” stands for “identify,” and “3typeA” means RNA’s “three types of modifications at adenosine sites.” Illustrated in Figure 3 is a flowchart to show the process of how the iRNA-3typeA predictor is working.
Figure 3.
A Flowchart to Show How the iRNA-3typeA Predictor Is Working
Cross-Validation
To evaluate the quality of a new predictor, we need to consider the following two problems. What metrics should be used to quantitatively display its performance? And what concrete procedure should be followed to derive the metrics’ values?
-
(1)
A set of four metrics. In literature, the following four conventional metrics are generally used to evaluate a predictor’s quality:77 (1) Acc, (2) MCC, (3) sensitivity (Sn), and (4) specificity (Sp). But the conventional expressions copied directly from math books are lacking in inductivity and hard to understand for most biological scientists. Fortunately, by using the symbols introduced by Chou in studying signal peptides,78 the four metrics can be converted to a set of intuitive ones58, 79 as given below:
| (Equation 6) |
where represents the total number of positive samples investigated, while is the number of positive samples incorrectly predicted to be negative, and represents the total number of negative samples investigated, while the number of the negative samples incorrectly predicted to be positive. With the set of formulations in Equation 6, the meanings of Sn, Sp, Acc, and MCC have become much more intuitive and easier to understand, as discussed in a series of recent studies in various biological areas (see, e.g., Liu et al.,21, 24, 28, 60 Ehsan et al.,26 Feng et al.,20, 27 Song et al.,31 Lin et al.,37 and Xu et al.80, 81).
-
(2)
Jackknife test. Now the next problem is how to test the values of these metrics in an objective way. As is well known, the independent dataset test, subsampling (or K-fold cross-validation) test, and jackknife test are the three cross-validation methods widely used for testing a prediction method.82 Of the three test methods, however, the jackknife test is deemed the least arbitrary and most objective one.32 Accordingly, the jackknife test has been widely recognized and increasingly adopted by investigators to examine the quality of various predictors (see, e.g., Ahmad et al.,52, 83 Lin et al.,84 Tang et al.,85 Tripathi and Pandey,86 and Dao et al.87). In view of this, the jackknife test was also adopted in the current study to examine the proposed predictor. During the jackknife test, each sample in the benchmark dataset is in turn singled out as an independent test sample and all the rule-parameters are calculated without including the one being identified. One more advantage of using the jackknife test is that there is no need to artificially separate the benchmark dataset into two subsets, one for training the model and one for testing it. This is because the outcome obtained by the jackknife test is actually a combination from many different independent dataset tests.88, 89, 90
Author Contributions
W.C. and H.L. designed the study; P.F., H.Y., and H.D. conducted the experiments; W.C., H.L., and K.-C.C. analyzed the results; W.C., H.L., and K.-C.C. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
The authors wish to thank the three anonymous reviewers, whose constructive comments were very helpful for further strengthening the presentation of this paper. This work was supported by the Natural Science Foundation of China (No. 31771471 and 61772119), the Natural Science Foundation for Distinguished Young Scholar of Hebei Province (No. C2017209244), the Program for the Top Young Innovative Talents of Higher Learning Institutions of Hebei Province (No. BJ2014028), and the Applied Basic Research Program of Sichuan Province (No. 2015JY0100).
Contributor Information
Wei Chen, Email: chenweiimu@gmail.com.
Hao Lin, Email: hlin@uestc.edu.cn.
References
- 1.Gilbert W.V., Bell T.A., Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016;352:1408–1412. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X., Xiong X., Wang K., Wang L., Shu X., Ma S., Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 2016;12:311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 8.Chen K., Lu Z., Wang X., Fu Y., Luo G.Z., Liu N., Han D., Dominissini D., Dai Q., Pan T., He C. High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew. Chem. Int. Ed. Engl. 2015;54:1587–1590. doi: 10.1002/anie.201410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helm M., Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 2017;18:275–291. doi: 10.1038/nrg.2016.169. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M., Pandolfi P.P. The epitranscriptome of noncoding RNAs in cancer. Cancer Discov. 2017;7:359–368. doi: 10.1158/2159-8290.CD-16-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nachtergaele S., He C. The emerging biology of RNA post-transcriptional modifications. RNA Biol. 2017;14:156–163. doi: 10.1080/15476286.2016.1267096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W., Tang H., Lin H. MethyRNA: a web server for identification of N6-methyladenosine sites. J. Biomol. Struct. Dyn. 2017;35:683–687. doi: 10.1080/07391102.2016.1157761. [DOI] [PubMed] [Google Scholar]
- 13.Qiu W.R., Jiang S.Y., Xu Z.C., Xiao X., Chou K.C. iRNAm5C-PseDNC: identifying RNA 5-methylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition. Oncotarget. 2017;8:41178–41188. doi: 10.18632/oncotarget.17104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W., Feng P., Yang H., Ding H., Lin H., Chou K.C. iRNA-AI: identifying the adenosine to inosine editing sites in RNA sequences. Oncotarget. 2017;8:4208–4217. doi: 10.18632/oncotarget.13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W., Feng P., Ding H., Lin H. PAI: predicting adenosine to inosine editing sites by using pseudo nucleotide compositions. Sci. Rep. 2016;6:35123. doi: 10.1038/srep35123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu W.R., Jiang S.Y., Sun B.Q., Xiao X., Cheng X., Chou K.C. iRNA-2methyl: identify RNA 2′-O-methylation sites by incorporating sequence-coupled effects into general PseKNC and ensemble classifier. Med. Chem. 2017;13:734–743. doi: 10.2174/1573406413666170623082245. [DOI] [PubMed] [Google Scholar]
- 17.Chen W., Tang H., Ye J., Lin H., Chou K.C. iRNA-PseU: identifying RNA pseudouridine sites. Mol. Ther. Nucleic Acids. 2016;5:e332. doi: 10.1038/mtna.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng P., Ding H., Chen W., Lin H. Identifying RNA 5-methylcytosine sites via pseudo nucleotide compositions. Mol. Biosyst. 2016;12:3307–3311. doi: 10.1039/c6mb00471g. [DOI] [PubMed] [Google Scholar]
- 19.Cheng X., Xiao X., Chou K.C. pLoc-mPlant: predict subcellular localization of multi-location plant proteins by incorporating the optimal GO information into general PseAAC. Mol. Biosyst. 2017;13:1722–1727. doi: 10.1039/c7mb00267j. [DOI] [PubMed] [Google Scholar]
- 20.Feng P., Ding H., Yang H., Chen W., Lin H., Chou K.C. iRNA-PseColl: identifying the occurrence sites of different RNA modifications by incorporating collective effects of nucleotides into PseKNC. Mol. Ther. Nucleic Acids. 2017;7:155–163. doi: 10.1016/j.omtn.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B., Wang S., Long R., Chou K.C. iRSpot-EL: identify recombination spots with an ensemble learning approach. Bioinformatics. 2017;33:35–41. doi: 10.1093/bioinformatics/btw539. [DOI] [PubMed] [Google Scholar]
- 22.Qiu W.R., Sun B.Q., Xiao X., Xu Z.C., Jia J.H., Chou K.C. iKcr-PseEns: identify lysine crotonylation sites in histone proteins with pseudo components and ensemble classifier. Genomics. 2017 doi: 10.1016/j.ygeno.2017.10.008. Published online November 16, 2017. [DOI] [PubMed] [Google Scholar]
- 23.Xiao X., Cheng X., Su S., Nao Q. pLoc-mGpos: Incorporate key gene ontology information into general PseAAC for predicting subcellular localization of Gram-positive bacterial proteins. Nat. Sci. 2017;9:331–349. [Google Scholar]
- 24.Liu L.M., Xu Y., Chou K.C. iPGK-PseAAC: identify lysine phosphoglycerylation sites in proteins by incorporating four different tiers of amino acid pairwise coupling information into the general PseAAC. Med. Chem. 2017;13:552–559. doi: 10.2174/1573406413666170515120507. [DOI] [PubMed] [Google Scholar]
- 25.Cheng X., Xiao X., Chou K.C. pLoc-mEuk: predict subcellular localization of multi-label eukaryotic proteins by extracting the key GO information into general PseAAC. Genomics. 2018;110:50–58. doi: 10.1016/j.ygeno.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Ehsan A., Mahmood K., Khan Y.D., Khan S.A., Chou K.C. A novel modeling in mathematical biology for classification of aignal peptides. Sci. Rep. 2018;8:1039. doi: 10.1038/s41598-018-19491-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng P., Yang H., Ding H., Lin H., Chen W., Chou K.C. iDNA6mA-PseKNC: identifying DNA N6-methyladenosine sites by incorporating nucleotide physicochemical properties into PseKNC. Genomics. 2018 doi: 10.1016/j.ygeno.2018.01.005. Published online January 31, 2018. [DOI] [PubMed] [Google Scholar]
- 28.Liu B., Yang F., Huang D.S., Chou K.C. iPromoter-2L: a two-layer predictor for identifying promoters and their types by multi-window-based PseKNC. Bioinformatics. 2018;34:33–40. doi: 10.1093/bioinformatics/btx579. [DOI] [PubMed] [Google Scholar]
- 29.Song J., Li F., Takemoto K., Haffari G., Akutsu T., Chou K.C., Webb G.I. PREvaIL, an integrative approach for inferring catalytic residues using sequence, structural, and network features in a machine-learning framework. J. Theor. Biol. 2018;443:125–137. doi: 10.1016/j.jtbi.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 30.Yang H., Qiu W.R., Liu G., Guo F.B., Lin H. iRSpot-Pse6NC: identifying recombination spots in Saccharomyces cerevisiae by incorporating hexamer composition into general PseKNC. Int. J. Biol. Sci. 2018 doi: 10.7150/ijbs.24616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song J., Wang Y., Li F., Akutsu T., Rawlings N.D. iProt-Sub: a comprehensive package for accurately mapping and predicting protease-specific substrates and cleavage sites. Brief. Bioinform. 2018 doi: 10.1093/bib/bby028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou K.C. Some remarks on protein attribute prediction and pseudo amino acid composition. J. Theor. Biol. 2011;273:236–247. doi: 10.1016/j.jtbi.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen H.B. Recent advances in developing web-servers for predicting protein attributes. Nat. Sci. 2009;1:63–92. [Google Scholar]
- 34.Cheng X., Zhao S.G., Lin W.Z., Xiao X., Chou K.C. pLoc-mAnimal: predict subcellular localization of animal proteins with both single and multiple sites. Bioinformatics. 2017;33:3524–3531. doi: 10.1093/bioinformatics/btx476. [DOI] [PubMed] [Google Scholar]
- 35.Cheng X., Xiao X., Chou K.C. pLoc-mGneg: predict subcellular localization of Gram-negative bacterial proteins by deep gene ontology learning via general PseAAC. Genomics. 2017 doi: 10.1016/j.ygeno.2017.10.002. Published online October 6, 2017. [DOI] [PubMed] [Google Scholar]
- 36.Cheng X., Xiao X., Chou K.C. pLoc-mHum: predict subcellular localization of multi-location human proteins via general PseAAC to winnow out the crucial GO information. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx711. Published online November 2, 2017. [DOI] [PubMed] [Google Scholar]
- 37.Lin H., Deng E.Z., Ding H., Chen W., Chou K.C. iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic Acids Res. 2014;42:12961–12972. doi: 10.1093/nar/gku1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia J., Liu Z., Xiao X., Liu B., Chou K.C. iPPI-Esml: an ensemble classifier for identifying the interactions of proteins by incorporating their physicochemical properties and wavelet transforms into PseAAC. J. Theor. Biol. 2015;377:47–56. doi: 10.1016/j.jtbi.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 39.Jia J., Zhang L., Liu Z., Xiao X., Chou K.C. pSumo-CD: predicting sumoylation sites in proteins with covariance discriminant algorithm by incorporating sequence-coupled effects into general PseAAC. Bioinformatics. 2016;32:3133–3141. doi: 10.1093/bioinformatics/btw387. [DOI] [PubMed] [Google Scholar]
- 40.Cheng X., Xiao X. pLoc-mVirus: predict subcellular localization of multi-location virus proteins via incorporating the optimal GO information into general PseAAC. Gene. 2017;628:315–321. doi: 10.1016/j.gene.2017.07.036. [DOI] [PubMed] [Google Scholar]
- 41.Chou K.C. Impacts of bioinformatics to medicinal chemistry. Med. Chem. 2015;11:218–234. doi: 10.2174/1573406411666141229162834. [DOI] [PubMed] [Google Scholar]
- 42.Chou K.C. An unprecedented revolution in medicinal chemistry driven by the progress of biological science. Curr. Top. Med. Chem. 2017;17:2337–2358. doi: 10.2174/1568026617666170414145508. [DOI] [PubMed] [Google Scholar]
- 43.Chen W., Feng P., Tang H., Ding H., Lin H. RAMPred: identifying the N(1)-methyladenosine sites in eukaryotic transcriptomes. Sci. Rep. 2016;6:31080. doi: 10.1038/srep31080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chou K.C. Prediction of protein cellular attributes using pseudo amino acid composition. Proteins. 2001;43:246–255. doi: 10.1002/prot.1035. [DOI] [PubMed] [Google Scholar]
- 45.Zhong W.Z., Zhou S.F. Molecular science for drug development and biomedicine. Int. J. Mol. Sci. 2014;15:20072–20078. doi: 10.3390/ijms151120072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou G.P., Zhong W.Z. Perspectives in medicinal chemistry. Curr. Top. Med. Chem. 2016;16:381–382. doi: 10.2174/156802661604151014114030. [DOI] [PubMed] [Google Scholar]
- 47.Esmaeili M., Mohabatkar H., Mohsenzadeh S. Using the concept of Chou’s pseudo amino acid composition for risk type prediction of human papillomaviruses. J. Theor. Biol. 2010;263:203–209. doi: 10.1016/j.jtbi.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 48.Mohabatkar H., Mohammad Beigi M., Esmaeili A. Prediction of GABAA receptor proteins using the concept of Chou’s pseudo-amino acid composition and support vector machine. J. Theor. Biol. 2011;281:18–23. doi: 10.1016/j.jtbi.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 49.Nanni L., Lumini A., Gupta D., Garg A. Identifying bacterial virulent proteins by fusing a set of classifiers based on variants of Chou’s pseudo amino acid composition and on evolutionary information. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2012;9:467–475. doi: 10.1109/TCBB.2011.117. [DOI] [PubMed] [Google Scholar]
- 50.Pacharawongsakda E., Theeramunkong T. Predict subcellular locations of singleplex and multiplex proteins by semi-supervised learning and dimension-reducing general mode of Chou’s PseAAC. IEEE Trans. Nanobioscience. 2013;12:311–320. doi: 10.1109/TNB.2013.2272014. [DOI] [PubMed] [Google Scholar]
- 51.Mondal S., Pai P.P. Chou’s pseudo amino acid composition improves sequence-based antifreeze protein prediction. J. Theor. Biol. 2014;356:30–35. doi: 10.1016/j.jtbi.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 52.Ahmad S., Kabir M., Hayat M. Identification of heat shock protein families and J-protein types by incorporating dipeptide composition into Chou’s general PseAAC. Comput. Methods Programs Biomed. 2015;122:165–174. doi: 10.1016/j.cmpb.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Kabir M., Hayat M. iRSpot-GAEnsC: identifing recombination spots via ensemble classifier and extending the concept of Chou’s PseAAC to formulate DNA samples. Mol. Genet. Genomics. 2016;291:285–296. doi: 10.1007/s00438-015-1108-5. [DOI] [PubMed] [Google Scholar]
- 54.Yu B., Li S., Qiu W.Y., Chen C., Chen R.X., Wang L., Wang M.H., Zhang Y. Accurate prediction of subcellular location of apoptosis proteins combining Chou’s PseAAC and PsePSSM based on wavelet denoising. Oncotarget. 2017;8:107640–107665. doi: 10.18632/oncotarget.22585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang S., Duan X. Prediction of protein subcellular localization with oversampling approach and Chou’s general PseAAC. J. Theor. Biol. 2018;437:239–250. doi: 10.1016/j.jtbi.2017.10.030. [DOI] [PubMed] [Google Scholar]
- 56.Muthu Krishnan S. Using Chou’s general PseAAC to analyze the evolutionary relationship of receptor associated proteins (RAP) with various folding patterns of protein domains. J. Theor. Biol. 2018;445:62–74. doi: 10.1016/j.jtbi.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 57.Chou K.C. Pseudo amino acid composition and its applications in bioinformatics, proteomics and system biology. Curr. Proteomics. 2009;6:262–274. [Google Scholar]
- 58.Chen W., Feng P.M., Lin H., Chou K.C. iRSpot-PseDNC: identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Res. 2013;41:e68. doi: 10.1093/nar/gks1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu W.R., Xiao X., Chou K.C. iRSpot-TNCPseAAC: identify recombination spots with trinucleotide composition and pseudo amino acid components. Int. J. Mol. Sci. 2014;15:1746–1766. doi: 10.3390/ijms15021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu B., Yang F., Chou K.C. 2L-piRNA: a two-layer ensemble classifier for identifying piwi-interacting RNAs and their function. Mol. Ther. Nucleic Acids. 2017;7:267–277. doi: 10.1016/j.omtn.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen W., Lei T.Y., Jin D.C., Lin H., Chou K.C. PseKNC: a flexible web server for generating pseudo K-tuple nucleotide composition. Anal. Biochem. 2014;456:53–60. doi: 10.1016/j.ab.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 62.Chen W., Zhang X., Brooker J., Lin H., Zhang L., Chou K.C. PseKNC-General: a cross-platform package for generating various modes of pseudo nucleotide compositions. Bioinformatics. 2015;31:119–120. doi: 10.1093/bioinformatics/btu602. [DOI] [PubMed] [Google Scholar]
- 63.Chen W., Lin H., Chou K.C. Pseudo nucleotide composition or PseKNC: an effective formulation for analyzing genomic sequences. Mol. Biosyst. 2015;11:2620–2634. doi: 10.1039/c5mb00155b. [DOI] [PubMed] [Google Scholar]
- 64.Chen W., Feng P., Tang H., Ding H., Lin H. Identifying 2′-O-methylationation sites by integrating nucleotide chemical properties and nucleotide compositions. Genomics. 2016;107:255–258. doi: 10.1016/j.ygeno.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 65.Li W.C., Deng E.Z., Ding H., Chen W., Lin H. iORI-PseKNC: a predictor for identifying origin of replication with pseudo k-tuple nucleotide composition. Chemometr. Intell. Lab. Syst. 2015;141:100–106. [Google Scholar]
- 66.Chou K.C. A vectorized sequence-coupling model for predicting HIV protease cleavage sites in proteins. J. Biol. Chem. 1993;268:16938–16948. [PubMed] [Google Scholar]
- 67.Feng P.M., Chen W., Lin H., Chou K.C. iHSP-PseRAAAC: identifying the heat shock protein families using pseudo reduced amino acid alphabet composition. Anal. Biochem. 2013;442:118–125. doi: 10.1016/j.ab.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 68.Feng P.M., Lin H., Chen W. Identification of antioxidants from sequence information using naïve Bayes. Comput. Math. Methods Med. 2013;2013:567529. doi: 10.1155/2013/567529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng P.M., Ding H., Chen W., Lin H. Naïve Bayes classifier with feature selection to identify phage virion proteins. Comput. Math. Methods Med. 2013;2013:530696. doi: 10.1155/2013/530696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen W., Feng P.M., Lin H., Chou K.C. iSS-PseDNC: identifying splicing sites using pseudo dinucleotide composition. BioMed Res. Int. 2014;2014:623149. doi: 10.1155/2014/623149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen W., Yang H., Feng P., Ding H., Lin H. iDNA4mC: identifying DNA N4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics. 2017;33:3518–3523. doi: 10.1093/bioinformatics/btx479. [DOI] [PubMed] [Google Scholar]
- 72.Chen X.X., Tang H., Li W.C., Wu H., Chen W., Ding H., Lin H. Identification of bacterial cell wall lyases via pseudo amino acid composition. BioMed Res. Int. 2016;2016:1654623. doi: 10.1155/2016/1654623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin H., Liang Z.Y., Tang H., Chen W. Identifying sigma70 promoters with novel pseudo nucleotide composition. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2017 doi: 10.1109/TCBB.2017.2666141. Published online February 8, 2017. [DOI] [PubMed] [Google Scholar]
- 74.Lai H.Y., Chen X.X., Chen W., Tang H., Lin H. Sequence-based predictive modeling to identify cancerlectins. Oncotarget. 2017;8:28169–28175. doi: 10.18632/oncotarget.15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao Y.W., Lai H.Y., Tang H., Chen W., Lin H. Prediction of phosphothreonine sites in human proteins by fusing different features. Sci. Rep. 2016;6:34817. doi: 10.1038/srep34817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang H., Tang H., Chen X.X., Zhang C.J., Zhu P.P., Ding H., Chen W., Lin H. Identification of secretory proteins in Mycobacterium tuberculosis using pseudo amino acid composition. BioMed Res. Int. 2016;2016:5413903. doi: 10.1155/2016/5413903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen J., Liu H., Yang J., Chou K.C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids. 2007;33:423–428. doi: 10.1007/s00726-006-0485-9. [DOI] [PubMed] [Google Scholar]
- 78.Chou K.C. Prediction of signal peptides using scaled window. Peptides. 2001;22:1973–1979. doi: 10.1016/s0196-9781(01)00540-x. [DOI] [PubMed] [Google Scholar]
- 79.Xu Y., Ding J., Wu L.Y., Chou K.C. iSNO-PseAAC: predict cysteine S-nitrosylation sites in proteins by incorporating position specific amino acid propensity into pseudo amino acid composition. PLoS ONE. 2013;8:e55844. doi: 10.1371/journal.pone.0055844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Y., Shao X.J., Wu L.Y., Deng N.Y., Chou K.C. iSNO-AAPair: incorporating amino acid pairwise coupling into PseAAC for predicting cysteine S-nitrosylation sites in proteins. PeerJ. 2013;1:e171. doi: 10.7717/peerj.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu Y., Wang Z., Li C., Chou K.C. iPreny-PseAAC: identify C-terminal cysteine prenylation sites in proteins by incorporating two tiers of sequence couplings into PseAAC. Med. Chem. 2017;13:544–551. doi: 10.2174/1573406413666170419150052. [DOI] [PubMed] [Google Scholar]
- 82.Chou K.C., Zhang C.T. Prediction of protein structural classes. Crit. Rev. Biochem. Mol. Biol. 1995;30:275–349. doi: 10.3109/10409239509083488. [DOI] [PubMed] [Google Scholar]
- 83.Ahmad K., Waris M., Hayat M. Prediction of protein submitochondrial locations by incorporating dipeptide composition into Chou’s general pseudo amino acid composition. J. Membr. Biol. 2016;249:293–304. doi: 10.1007/s00232-015-9868-8. [DOI] [PubMed] [Google Scholar]
- 84.Lin H., Liu W.X., He J., Liu X.H., Ding H., Chen W. Predicting cancerlectins by the optimal g-gap dipeptides. Sci. Rep. 2015;5:16964. doi: 10.1038/srep16964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang H., Zou P., Zhang C., Chen R., Chen W., Lin H. Identification of apolipoprotein using feature selection technique. Sci. Rep. 2016;6:30441. doi: 10.1038/srep30441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tripathi P., Pandey P.N. A novel alignment-free method to classify protein folding types by combining spectral graph clustering with Chou’s pseudo amino acid composition. J. Theor. Biol. 2017;424:49–54. doi: 10.1016/j.jtbi.2017.04.027. [DOI] [PubMed] [Google Scholar]
- 87.Dao F.Y., Yang H., Su Z.D., Yang W., Wu Y., Hui D., Chen W., Tang H., Lin H. Recent advances in conotoxin classification by using machine learning methods. Molecules. 2017;22:e1057. doi: 10.3390/molecules22071057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chou K.C., Shen H.B. Recent progress in protein subcellular location prediction. Anal. Biochem. 2007;370:1–16. doi: 10.1016/j.ab.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 89.Chou K.C., Shen H.B. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008;3:153–162. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- 90.Shen H.B. Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010;2:1090–1103. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- 91.Frank E., Hall M., Trigg L., Holmes G., Witten I.H. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20:2479–2481. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]