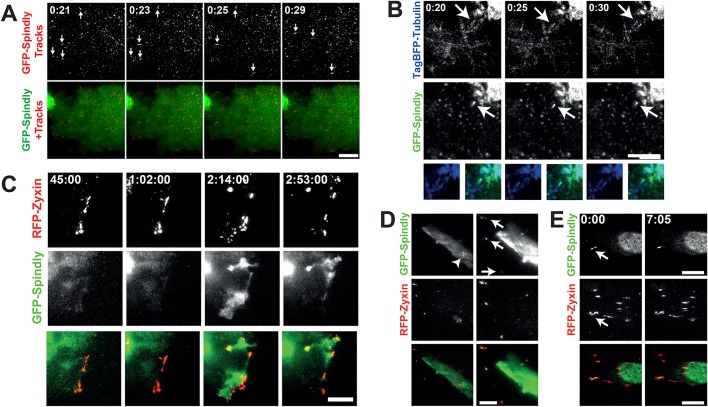
Fig. 2.
Spindly can be seen moving on the basal cell cortex and associated with microtubule plus-ends. (A) U2OS cells stably expressing low levels of GFP-Spindly were imaged with total internal reflection fluorescence microscopy (TIRF). To better visualize the movement of individual particles across multiple images, a Gaussian filter was applied to de-noise the images, and then a temporal filter was used to find foci that were moving across multiple images that were compiled into trails. Arrows show moving GFP-Spindly. The lower image shows the particle trails (red) overlaid on top of the still GFP-Spindly image. Time is minutes:seconds. (B) The same U2OS cells as in A were transfected with TagBFP2-Tubulin and imaged in TIRF. The arrow shows a TagBFP2-labeled microtubule that shrinks and grows over the course of imaging. Time is minutes:seconds. (C) The same U2OS cells were transfected with RFP-Zyxin, grown to confluency and then induced to migrate. GFP-Spindly only began to co-localize at the leading edge at 45 min post scratch-wounding. Time is shown in hours:minutes:seconds. (D) Fibroblasts were transiently transfected with GFP-Spindly and RFP-Zyxin and then imaged in TIRF. Arrows show focal adhesions where Spindly and Zyxin colocalize, which are more obvious in the first frame where the brightness was enhanced relative to the other time points. The arrowhead shows filaments of GFP-Spindly. (E) In another fibroblast, a focus of GFP-Spindly was observed (arrow) that was dynamically associated with focal adhesions. Time is shown in minutes:seconds. Scale bars: 10 µm.