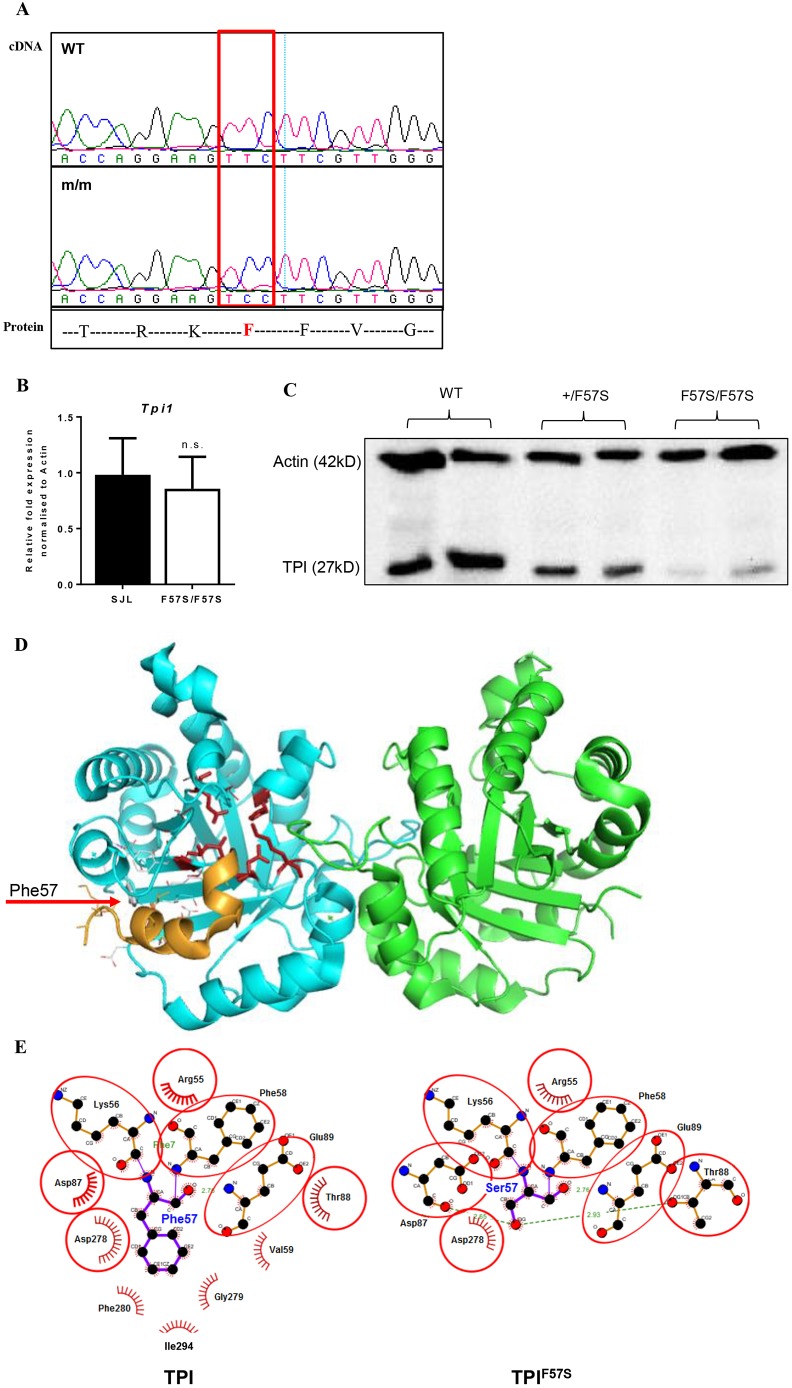
Fig. 2.
Characterisation of the F57S mutation. (A) Sanger sequencing of RBC19 homozygous cDNA shows a T→C substitution in Tpi1, resulting in a phenylalanine (F) to serine (S) amino acid change. (B) Quantitative RT-PCR for relative Tpi1 mRNA expression in the bone marrow of wild-type (WT) and homozygous (F57S/F57S) mice. Data are mean±s.d.; n=4. (C) Western blot of the TPI protein from bone marrow lysates of WT, heterozygous (+/F57S) and homozygous (F57S/F57S) mice. (D) 3D protein modelling of the murine TPI dimer in ribbons (blue and green), showing the enzyme active site (red sticks). The location of F57 is highlighted (red arrow to grey stick), as well as its adjacent binding helix (orange ribbon). (E) 2D contact map of the F57 site and its surrounding amino acids in both the WT (TPI) and mutant (TPIF57S) forms. In the mutant, the F57S substitution is predicted to distort surrounding amino acid interactions and subsequently induce conformational changes to the protein.