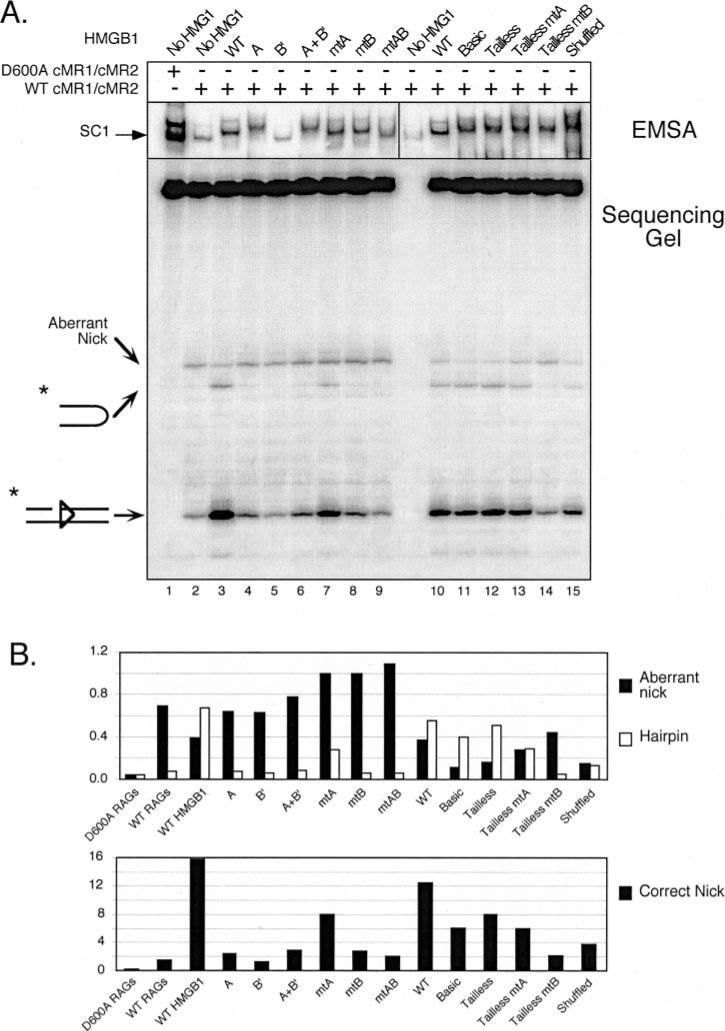
Fig. 4. Stimulation of RAG-mediated 23-RSS cleavage by HMGB1 is primarily mediated by box B, but requires a tethered box A.
A, in-gel cleavage assay of RAG-RSS complexes super-shifted by different forms of HMGB1. A wild-type or inactive RAG complex (WT cMR1/cMR2 and D600A cMR1/cMR2, respectively) was incubated with radiolabeled intact 23-RSS either alone or with various forms of HMGB1 (indicated above the gel) in preparative binding reactions containing Ca2+ and protein-DNA complexes were fractionated by EMSA. After electrophoresis, gels were submerged in buffer containing Mg2+ for 1 h at 37 °C to initiate cleavage and then the DNA was transferred to DEAE cellulose paper. Reaction products recovered from complexes of interest (identified by autoradiography; see exposure at the top) were then analyzed on a 15% polyacrylamide sequencing gel (bottom). Positions of nicked, hairpin, and aberrantly nicked products are indicated at the left. Note that samples from lanes 1–9 and 10 –15 were obtained from different in-gel cleavage assays (given space limitations for the EMSA), but all reaction products were fractionated on the same sequencing gel. B, quantification of reaction products. The abundance of aberrant nicks and hairpins (top bar graph) or correctly positioned nicks (bottom bar graph) are shown for each lane depicted in A and presented in the same order. The data are representative of independent experiments.