Figure 3.
CRISPR/Cas9 Mediates Disruption of APPSW or APPWT Allele in Patient-Derived Fibroblasts
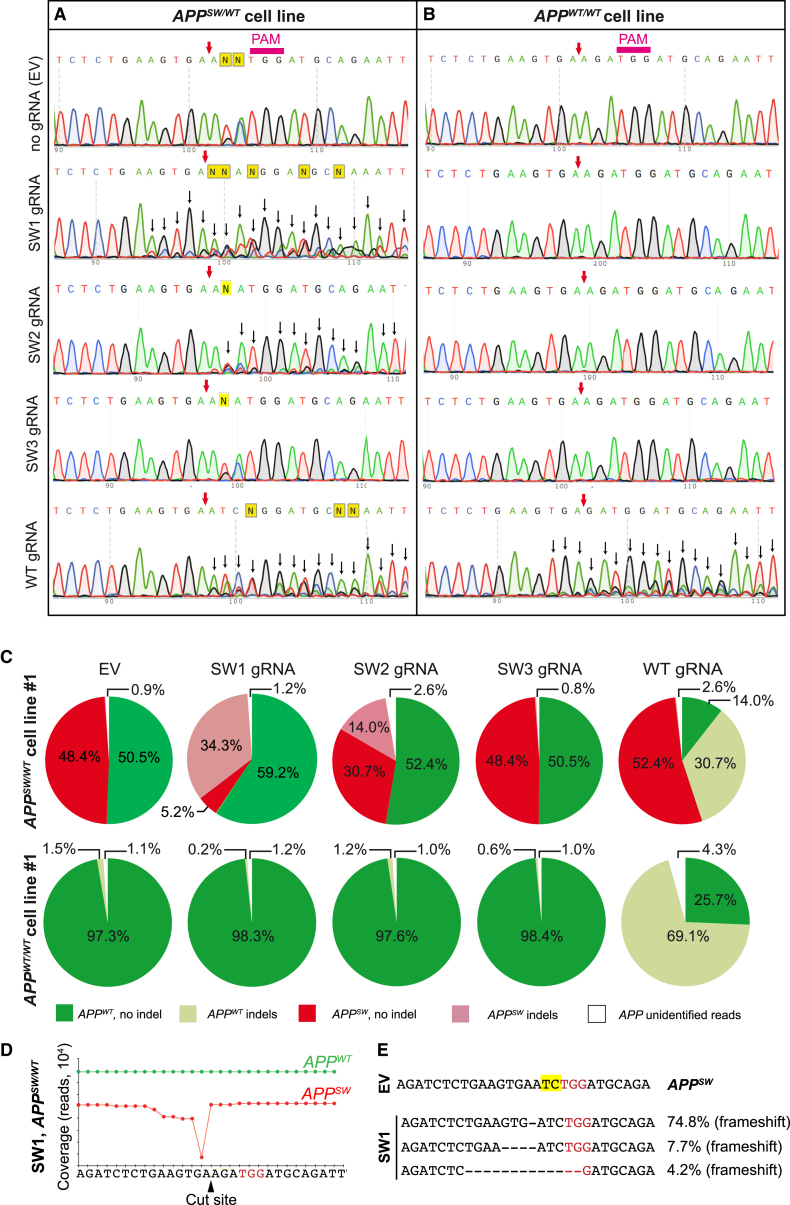
(A and B) Sanger sequencing of the APP gene from mutation carrier No. 1 and control line No. 1 transfected with Cas9-2A-GFP and gRNA targeting the APPSW (SW1, SW2, and SW3) or APPWT allele (WT), respectively. GFP-expressing cells were sorted 3 days after transfection. On the APPSW/WT cell line (A), SW1, SW2, and WT gRNA were disruptive and created indels (as indicated by background peaks [i.e., heterogeneity in reads] downstream of the cut site, black arrows). On the APPWT/WT cell line (B), WT gRNA led to indel formation, whereas SW1, SW2, or SW3 gRNA did not. PAM, protospacer-adjacent motif. The purple arrow shows the predicted CRISPR cut site. (C) Targeted deep sequencing of the APP allele from CRISPR-treated APPSW/WT fibroblasts (top pie charts) and control APPWT/WT fibroblasts (bottom pie charts) is shown. Colors indicate mutant (red), wild-type (green), mutant with indels (light red), and wild-type with indels (light green) reads. For some reads, we were not able to determine their origins as mutant or wild-type (due to sequencing errors or larger deletions), and these are marked as white areas on the pie charts. SW1 and SW2 gRNA resulted in indel formation in the APPSW, but not in the APPWT allele. WT gRNA resulted in indel formation only in the APPWT allele in APPSW/WT cells and APPWT/WT cells. SW3 did not lead to indel formation in any of the alleles. (D) Coverage of the APP allele in APPSW/WT cells, treated with Cas9 and SW1 gRNA, is shown. Deletions were detectable adjacent to the CRISPR cut site on the APPSW allele (red trace). No deletions were present on the APPWT allele. The PAM site is marked in red. (E) The most frequent reads in the APPSW/WT fibroblasts treated with Cas9 and SW1 gRNA are shown. PAM sites are marked with red, and the APPswe mutation is marked in yellow. The percentages refer to the fraction of reads that contained any indels.