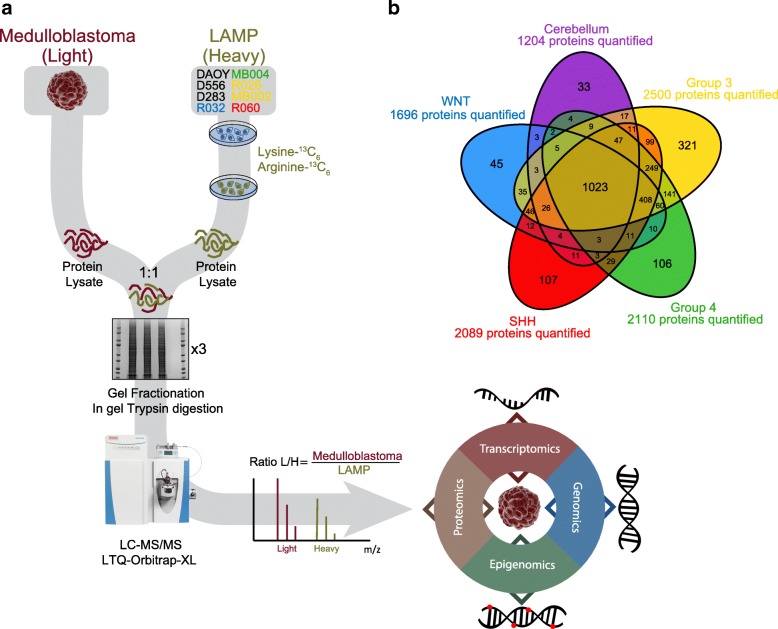
Fig. 1.
SILAC proteomics workflow and output. a The Labeled Atlas of Medulloblastoma Proteins (LAMP) was generated by combining equal amounts of isotopically labeled (Lysine-13C6, and Ariginine13C6) proteins from 8 primary and established cell lines, representing the four primary medulloblastoma subgroups. Protein lysates from tissues were spiked 1:1 with the SILAC reference atlas (LAMP). The resulting protein lysates were fractionated on a 1-D gel in triplicate, trypsin digested, and further fractionated by HPLC in line with the MS/MS analysis. The spectra were then searched against the Uniprot or custom protein databases to identify peptide sequences and their originating proteins. Protein quantitation is derived from the ratio of the light tissue peptides relative to the heavy SILAC reference peptides. The proteomic data was functionally integrated with tumor-matched transcriptomic, epigenomic, and genomic data. b Venn diagram of quantified proteins along the 4 medulloblastoma subgroups and cerebellum tissues. A total of 2901 proteins were quantified