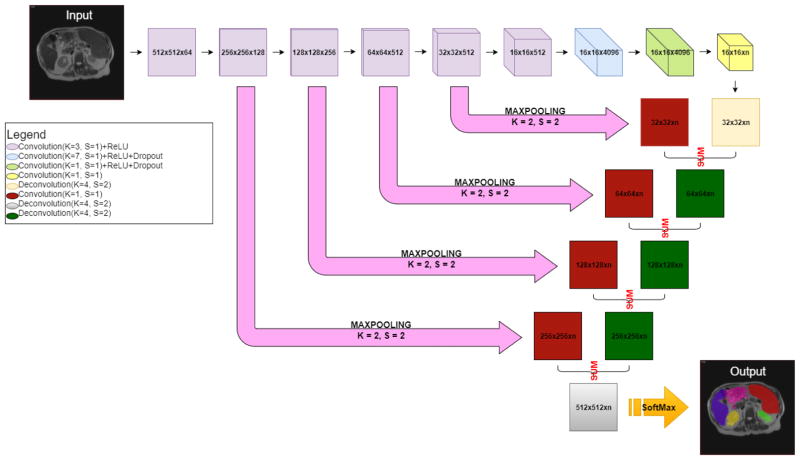
Figure 2.
The structure of the FCNN. The encoder is shown as the top row, where the resolution and number of channels of the output feature maps for each convolutional layer are shown in each box. For instance, the “512 × 512 × 64” means the resolution is 512 × 512 and the number of channels is 64. Then the decoder units performed the deconvolutional operations to expand the feature maps from 16 × 16 resolution to 512 × 512 resolution. To propagate the spatial information of the feature maps, the skip connection layers performed the convolutional operations to convert the number of the channels to two. The convolved feature maps were added with the deconvolved feature maps from the decoder. Finally, we derived the predications with the same size as the input image. “n” is the number of output channels (number of organs + background). The ReLU activate function was used in the activation layers. The dropout layers were used to alleviate the overfitting issue. The 2D cross-entropy loss was used as the loss function. The output hard segmentations were derived from channel number with maximum intensities on the heatmaps after the softmax layers. Specifically, our FCNN is structurally identical to the one used in [1].