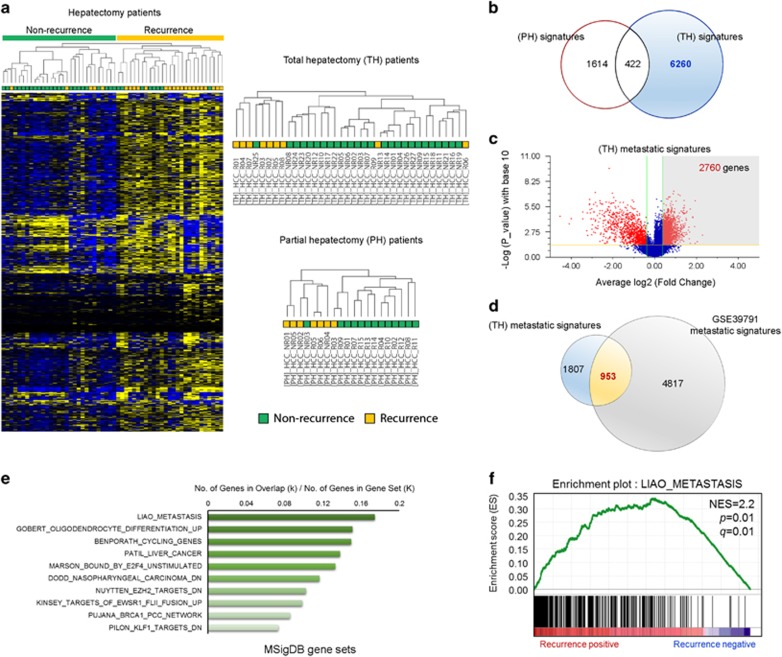
Figure 1.
Identification and selection of genes associated with the recurrence of hepatocellular carcinoma (HCC). (a) Two-dimensional diagram of the differential expression of 7550 genes; the data were organized according to non-recurrence or recurrence HCC tissues from patients who underwent liver resection. Dendrograms were derived from clustering the data for patients with total hepatectomy (TH) or partial hepatectomy (PH). Blue and yellow colors indicate downregulation and upregulation, respectively. (b) Venn diagram of genes altered between the TH and PH groups. (c) Volcano plot of differentially upregulated genes between non-recurrence and recurrence groups in patient with total hepatectomy. (d) The 953 common upregulated genes between the metastatic signature from patients with TH and that from the GSE39791 data set. (e) Bar chart of the top 10 genes among the 953 genes identified in the overt metastatic molecular signature using MSigDB. (f) Gene Set Enrichment Analysis (GSEA) was performed with the recurrence-associated molecular signature and LIAO_METASTASIS gene set. The barcode indicates gene positions. The y axis indicates the extent of enrichment.