Abstract
Background
Toll-like receptors (TLRs) play essential role in innate and acquired immunity, are expressed in various cell types, and are associated with altered susceptibility to many diseases, and cancers. The aim of this study was to investigate TLR2 (-196 to-174del), TLR4 (Asp299Gly and Thr399Ile) and TLR9 (T1237C and T1486C) gene polymorphisms at risk of colorectal cancer (CRC) development and progression.
Methods
Peripheral blood was obtained from 397 patients with adjuvant (stage II/III, n = 202) and metastatic (n = 195) CRC. Moreover, blood samples from 50 healthy volunteers and 40 patients with adenomatous polyps were also included as control groups. DNA from patients and controls was analyzed using PCR and PCR-RFLP for genotyping functional polymorphism within TLR2, TLR4 and TLR9 genotypes.
Results
TLR2–196 to-174del/del genotype was detected in 76.6% of the patients and was significantly higher that the controls groups (p<0.001). TLR4 Asp299Gly, TLR4 Thr399Ile, TLR9 T1237C and T1486C homozygous genotypes were detected in 70.5%, 70.5%, 61.5% and 61.5% of the patients respectively, and were also significantly higher than that in the control groups (p<0.001). All polymorphisms detected were also significantly associated with the metastatic disease (p<0.001) leading to shorter overall survival (p<0.001); whereas, TLR4 Asp299Gly and Thr399Ile polymorphisms were significantly associated with KRAS mutations.
Conclusions
The detection of higher frequencies of the TLR2, TLR4 and/or TLR9 polymorphisms in CRC patients compared with the control groups highlight the role of these polymorphism in CRC development and cancer progression.
Introduction
Colorectal cancer (CRC) represents the 9% of all malignancies and is considered as an important cause of death worldwide [1]. The development of CRC has been associated with genetic mutations, diet, inflammatory processes and the gut microflora, with the 95% of CRC cases been characterized as sporadic cancer [2]. The relationship between cancer and the microorganisms has been shown in various organs, with the most known example being the relation between H. pylori and gastric cancer [3].
Toll-like receptors (TLRs) play an essential role in both innate and acquired immunity and are expressed in various types of cells, including cancer cells [4–6]. Several polymorphisms within the TLR genes are associated with altered susceptibility to many diseases (infections, allergies, inflammatory diseases) as well as cancers [7] and TLR signaling plays a role in promoting malignant cell survival [8].
CRC development and progression have been correlated with TLR2 and TLR4 overexpression [9–10]. In fact, variant -196 to -174 chromosome 4q32 22-bp insertion/deletion (Ins/Del) polymorphisms in the promoter region, alters TLR2 promoter activity and thus its level of expression [11]. Moreover, TLR4 is expressed in CRC and promotes cancer cells to escape immune-surveillance by stimulating immunosuppressive agents and resistance to apoptosis [12]. Two common mutations, Asp299Gly and Thr399Ile, which occur in exon 4 of the human TLR4 gene (A896G and C1196T, respectively), are located within the extracellular domain of the receptor [13], have been associated with increased risk to Crohn’s disease and ulcerative colitis [14–16]. Additionally, human TLR9 occurs at 3p21.3, a region frequently deleted in human cancers [17]. Among the studied polymorphisms, variants T1237 and T1486C have been associated with the risks of multiple tumors; however, due to limited sample sizes, the reported results are inconsistent.
Herein, we hypothesized that the presence of TLR2, TLR4 and TLR9 variants affect gut homeostasis resulting in impairment of TLRs activation, thus leading to inflammation and CRC development and progression. Thus, we aimed to a) evaluate the expression of -196 to -174 del allele in the TLR2 gene, the Asp299Gly and Thr399Ile polymorphisms in the TLR4 gene and the T1237 and T1486C polymorphisms of the TLR9 gene in patients with early CRC; b) compare their expression in patients with metastatic disease; c) determine whether patients carrying TLR alleles have an increased risk of recurrence and decreased overall survival and d) associate TLR polymorphisms with patients’ molecular profiling. These results are expected to show whether patients carrying TLR alleles have an increased risk of recurrence and decreased overall survival. Finally, the understanding of how TLRs enhance angiogenesis could help improve the development of anticancer agents and targeted therapies.
Materials and methods
Patients’ population
Since September 2003 to November 2013, 397 consecutive patients with newly diagnosed colon adenocarcinoma, treated at the Department of Medical Oncology, University Hospital of Heraklion were enrolled in the study. Disease status was coded, without the knowledge of the laboratory analysis. The study was approved by the Ethics Committee/Institutional review board of the University Hospital of Heraklion and signed informed consent has been obtained from all enrolled patients.
Blood and tissue samples from control groups
Peripheral blood (15 ml in EDTA) was obtained from 50 healthy blood donors and was used as controls in the study. Moreover, formalin-fixed paraffin embedded (FFPE) tissues from 40 patients with colon adenomas, in the absence of CRC, were used as extra controls in the study.
Genomic DNA extraction
Peripheral blood mononuclear cells (PBMC) from patients and healthy blood donors were obtained by Ficoll–Hypaque density gradient (d = 1,077 g/ml; Sigma-Aldrich, GmbH, Germany) centrifugation at 1,800 rpm for 30 min.
Representative formalin-fixed, paraffin-embedded (FFPE) primary tumor specimens were reviewed by an experienced pathologist in order to ensure the validity of the specimen and define the most appropriate area for microdissection. Malignant cells were procured using a piezoelectric micro dissector (Eppendorf, Germany) as previously described [18].
DNA extraction both from blood and tissue samples was performed using the MasterPure™ Complete DNA and RNA Purification Kit (Epicenter, Madison, Wisconsin, USA) following the manufacturer’s instructions. DNA was quantified using the NanoDrop ND-1000 v3.3 (ThermoFisher Scientific, Waltham, Massachusetts, USA) equipment and the samples were stored at -20°C until their use.
TLR genotyping
Polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) was performed for genotyping of single nucleotide substitutions of the TLR2, TLR4 and TLR9 genes. Primers used for PCR amplification of the fragments are shown in S1 Table. In brief, determination of TLR2–196 to -174 Ins/Del polymorphism was performed by PCR as previously described [19]; while TLR4 (Asp299Gly and Thr399Ile) and TLR9 (T1237 and T1486C) genotyping were determined by PCR-RFLP. Allele types, single nucleotide polymorphism reference numbers and PCR conditions for all the analyzed polymorphisms are shown in S2 Table.
The amplified products of the TLR4 (Asp299Gly and Thr399Ile) and TLR9 (T1237 and T1486C) genes were digested using restriction enzymes; NcoI (ThermoFisher Scientific), HinfI (New England BioLabs, United Kingdom), BstNI (New England BioLabs) and AflII (New England BioLabs), respectively, according to manufacturer’s instructions. Briefly, 10 μl of each related PCR product was mixed with 1.5 μl of each restriction enzyme, 2 μl of 10× buffers and 7 μl H2O. After incubation at 37°C for 16 h, the restriction fragments were separated by electrophoresis on a 3% agarose gel, stained with Sybr Safe DNA Gel Stain (ThermoFisher Scientific), and visualized with the AlphaImager ultraviolet transilluminator (Alpha Innotech Corp., San Leandro, CA).
Study design and statistics
This is a retrospective, single institution study, investigating the TLR2, TLR4 and TLR9 gene polymorphisms in PBMCs from patients with CRC before the initiation of adjuvant or first-line treatment. Disease-free survival (DFS) was calculated from the date of surgery to the date of disease recurrence, diagnosis of new colorectal primary or death from any cause, while progression-free survival (PFS) was calculated from the day of diagnosis to documented disease progression or death from any cause. Overall survival (OS) was calculated from the day of diagnosis to the date of death, from any cause. Laboratory analysis was carried out blindly to clinical data. Statistical analysis was based on contingency tables, including calculations of hazard rations (HR) and 95% CI. The two-tailed Fisher’s exact test was applied to determine whether distributions of categorized variables between the groups were significant. The association of risk factors with time-to-event endpoints was analyzed with the log rank test and the Kaplan–Meier method was used to plot the corresponding DFS, PFS and OS curves. The Hardy–Weinberg equilibrium was tested by comparing expected and observed genotype frequencies by chi-square test. Statistical significance was set at p = 0.05.
Results
Patients’ demographics and molecular characteristics
From 09/2013 to 11/2013, 397 newly diagnosed and histologically documented patients with CRC were enrolled in the study. The patients’ characteristics are listed in Table 1. The median age was 65 years, 246 (62.0%) patients were males, 202 (50.9%) was of stage IIA-IIIC, 279 (70.3%) had a colon/sigmoid tumor location, 372 (93.7%) had PS (ECOG) 0–1, and 205 (47.4%) had a high grade tumors. Moreover, 347 (87.8%) and 80 (20.4%) patients underwent surgery and radiotherapy, respectively; 230 (58.7%) patients received an adjuvant treatment, 64 (27.8%) of which were relapsed, whereas 223 (56.2%) patients received front-line treatment, 197 (88.3%) of which relapsed (Table 1 and S3 Table).
Table 1. Patients’ demographics and tumor’s characteristics.
| Frequency | % | |
|---|---|---|
| Age (range) | 65 (18–88) | |
| <70 | 260 | 65,5 |
| > = 70 | 137 | 34,5 |
| Gender | ||
| Male | 246 | 62,0 |
| Female | 151 | 38,0 |
| Stage | ||
| IIA-IIIC | 202 | 50,9 |
| IV | 195 | 49,1 |
| Location | ||
| Colon/Sigmoid | 279 | 70,3 |
| Rectum | 118 | 29,7 |
| PS (ECOG) | ||
| 0–1 | 372 | 93,7 |
| > = 2 | 25 | 6,3 |
| Radiotherapy | ||
| Yes | 80 | 20,4 |
| No | 313 | 79,6 |
| Adjuvant Treatment | ||
| Yes | 230 | 58,7 |
| No | 162 | 41,4 |
| First Line Treatment | ||
| Yes | 223 | 56,2 |
| No | 174 | 46,8 |
| Grade | ||
| High | 205 | 47,4 |
| Low | 228 | 52.6 |
| TLR2 196-to -174 del | ||
| ins/del | 93 | 23,4 |
| del/del | 304 | 76,6 |
| TLR4 Asp299Gly (A/G) | ||
| AA | 11 | 2,8 |
| AG | 106 | 26,7 |
| GG | 280 | 70,5 |
| TLR4 Thr399Ile (T/C) | ||
| CC | 11 | 2,8 |
| CT | 106 | 26,7 |
| TT | 280 | 70,5 |
| TLR9 T1237C | ||
| TC | 153 | 38,5 |
| CC | 244 | 61,5 |
| TLR9 T1486C | ||
| TC | 153 | 38,5 |
| CC | 244 | 61,5 |
| KRAS | ||
| Mutant | 104 | 42,4 |
| Wt | 141 | 57,6 |
| ND | 152 |
Analysis of the TLR2 gene (-196 to -174 del) polymorphisms
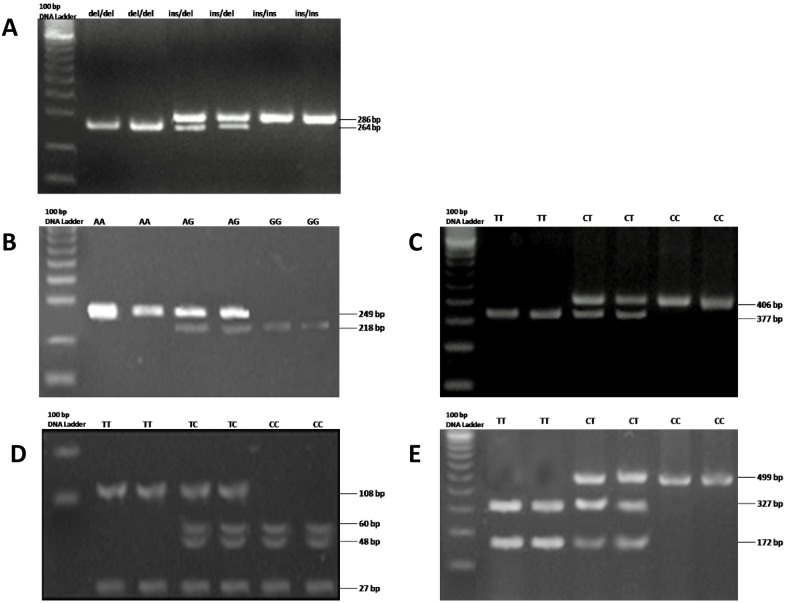
Determination of the TLR2 genotype was based on the presence of the specific band (Fig 1a). As indicated by the PCR gel electrophoresis, 93 (23.4%) patients presented heterozygous (ins/del) alleles, by simultaneously presenting the 286 bp 218 bp bands; whereas 304 (76.6%) patients presented the homozygous mutant (del/del) allele, by amplifying an 286 bp band (Table 1 and Fig 1a). Compared with the healthy donors and the patients with adenomatous polyps control groups, there was a significant association (p<0.001) in genotype and allelic frequency, since both control groups presented only the homozygous for the wild type (ins/ins) allele, by amplifying only the 286 bp band (Table 2, Fig 1a and S3 Table).
Fig 1. Representative agarose gels showing PCR-RFLP analysis of different single nucleotide polymorphisms of the a) TLR2–196 to 174 del, b) TLR4 Asp299Gly, c) TLR4 Thr399Ile, d) TLR9 T1276C and e)TLR4 T1486C, respectively.
Table 2. Association of patients’ and control groups TLR gene polymorphisms.
| Gene | SNP | Genotype | Patient No (%) | Healthy blood donors No (%) | Adenomatous polyps No (%) | p value |
|---|---|---|---|---|---|---|
| TLR2 | -196 to -174 bp | ins/ins | 0 (0,0) | 50 (100,0) | 40 (100,0) | <0.001 |
| ins/del | 93 (23,4) | 0 (0,0) | 0 (0,0) | |||
| del/del | 304 (76,6) | 0 (0,0) | 0 (0,0) | |||
| TLR4 | Asp299Gly (A/G) | Wt | 11 (2,8) | 50 (100,0) | 36 (90,0) | <0.001 |
| Hetero | 106 (26,7) | 0 (0,0) | 4 (10,0) | |||
| Homo | 280 (70,5) | 0 (0,0) | 0 (0,0) | |||
| Thr399Ile (T/C) | Wt | 11 (2,8) | 50 (100,0) | 36 (90,0) | <0.001 | |
| Hetero | 106 (26,7) | 0 (0,0) | 4 (10,0) | |||
| Homo | 280 (70,5) | 0 (0,0) | 0 (0,0) | |||
| TLR9 | T1237C | Wt | 0 (0,0) | 26 (52,0) | 10 (25,0) | <0.001 |
| Hetero | 153 (38,5) | 24 (48,0) | 30 (75,0) | |||
| Homo | 244 (61,5) | 0 (0,0) | 0 (0,0) | |||
| T1486C | Wt | 0 (0,0) | 26 (52,0) | 10 (25,0) | <0.001 | |
| Hetero | 153 (38,5) | 24 (48,0) | 30 (75,0) | |||
| Homo | 244 (61,5) | 0 (0,0) | 0 (0,0) |
Analysis of the TLR4 gene (Asp299Gly and TLR4 Thr399Ile) polymorphisms
The expected amplification products of the TLR4 gene Asp299Gly (A/G) and Thr399Ile (T/C) polymorphisms were 249 bp and 406 bp, respectively. The PCR products, for Asp299Gly (A/G) and Thr399Ile (T/C) polymorphisms were digested by NcoI and HinfI enzymes, respectively. Following electrophoresis, 280 (70.5%), 106 (26.7%) and 11 (2.8%) patients presented the homozygous mutant, the heterozygous and the homozygous wild type genotype, for both polymorphisms (Table 1 and Fig 1b and 1c). Compared with the control groups, there was a significant association (p<0.001) in genotype and allelic frequency of both polymorphisms, since the healthy donors control group presented only the homozygous wild type genotype. Moreover, 36 (90%) and 4 (10%) adenomatous polyps control patients presented the homozygous wild type and the heterozygous polymorphisms, respectively, and this was observed both in the cases of Asp299Gly (A/G) and Thr399Ile (T/C) polymorphisms (Table 2, Fig 1b and 1c and S3 Table).
Analysis of the TLR9 gene (T1237C and T1438C) polymorphisms
The expected amplification product of the TLR9 gene T1237C polymorphism was 108 bp and 27 bp; whereas the length of the specific ampification band of the T1486C polymorphism was 499 bp. The PCR products, for T1237C and T1486C polymorphisms were digested by AflII and BstNI enzymes, respectively. Following electrophoresis, 244 (61.5%) and 153 (38.5%) patients presented the homozygous mutant and the heterozygous genotype, for both polymorphisms. Not one of the patients presented the homozygous wild type genotype. (Table 1 and Fig 1d and 1e). Compared with the control groups, there was a significant association (p<0.001) in genotype and allelic frequency of both polymorphisms, since 26 (52%) and 24 (48%) healthy donors presented only the homozygous wild type and the heterozygous genotype, respectively; whereas, 10 (25%) and 30 (75%) adenomatous polyps control patients presented the homozygous wild type and the heterozygous polymorphisms, respectively, and this was observed in both T1237C and T1486C polymorphisms (Table 2, Fig 1d and 1e and S3 Table).
Association of TLR2, TLR4 and TLR9 variants and disease stage
The association between the TLR variants analyzed and the patients’ disease stage is presented in Table 3. The TLR2 homozygous mutant (del/del) allele was more prevalent in stage IV patients whereas, the heterozygous (ins/del) allele was mostly seen in stage IIA-IIIC patients (95.9% vs 57.9% and 4.1% vs 42.1%, respectively; p<0.001) (Table 3). The TLR4 Asp299Gly (A/G) homozygous mutant genotype was more prevalent in stage IV patients, whereas the heterozygous and homozygous wild type genotype were mostly prevalent in stage IIA-IIIC patients (84.1% vs 57.1%, 13.8% vs 39.1% and 2.1% vs 3.5%, respectively; p<0.001); and this was also observed in the case of Thr399Ile (T/C) polymorphism (Table 3). Moreover, the TLR9 T1237C and T1486C homozygous mutant genotypes were also more prevalent in stage IV patients, whereas the heterozygous genotype was mostly met in stage IIA-IIIC patients (73.3% vs 50.0% and 26.7% vs 50.0%, respectively; p<0.001) (Table 3 and S3 Table).
Table 3. Association of TLR2, TLR4 and TLR9 variants and patients’ disease stage.
| Gene | SNP | Genotype | IIA-IIIC | IV | p-value |
|---|---|---|---|---|---|
| TLR2 | -196 to -174 bp | ins/del | 85 (42,1%) | 8 (4,1%) | <0,001 |
| del/del | 117 (57,9%) | 187 (95,9%) | |||
| TLR4 | Asp299Gly (A/G) | WT | 7 (3,5%) | 4 (2,1%) | <0,001 |
| Hetero | 79 (39,1%) | 27 (13,8%) | |||
| Homo | 116 (57,4%) | 164 (84,1%) | |||
| Thr399Ile (T/C) | WT | 7 (3,5%) | 4 (2,1%) | <0,001 | |
| Hetero | 79 (39,1%) | 27 (13,8%) | |||
| Homo | 116 (57,4%) | 164 (84,1%) | |||
| TLR9 | T1237C | Hetero | 101 (50,0%) | 52 (26,7%) | <0,001 |
| Homo | 101 (50,0%) | 143 (73,3%) | |||
| T1486C | Hetero | 101 (50,0%) | 52 (26,7%) | <0,001 | |
| Homo | 101 (50,0%) | 143 (73,3%) |
Association of TLR2, TLR4 and TLR9 variants and KRAS status
Table 4 shows the association of the TLR variants according to the patients’ KRAS status. The TLR2 homozygous mutant (del/del) allele was more prevalent in patients with a wild type KRAS status whereas the heterozygous (ins/del) allele was mostly seen in KRAS mutants (86.5% vs 77.9% and 13.5% vs 22.1%, respectively), although not significant (p = 0.076). Asp299Gly (A/G) and Thr399Ile (T/C) TLR4 homozygous mutant genotype were more frequent in KRAS mutants, whereas heterozygous genotypes were more prevalent in KRAS wild type patients (80.8% vs 70.2% and 15.4% vs 28.4%, respectively; p = 0.028). On the contrary, both TLR9 T1237C and T1486C homozygous mutant genotypes were more prevalent in KRAS mutants, whereas the heterozygous genotypes were more frequent in KRAS wild type patients, although not significant (68.3% vs 58.2% and 31.7% vs 41.8%, respectively; p = 0.106) (Table 4 and S3 Table).
Table 4. Association of TLR2, TLR4 and TLR9 variants according to KRAS status.
| KRAS | |||||
|---|---|---|---|---|---|
| Gene | SNP | Genotype | Wt | Mutant | p value |
| TLR2 | -196 to -174 bp | ins/del | 19 (13,5%) | 23 (22,1%) | 0,076 |
| del/del | 122 (86,5%) | 81 (77,9%) | |||
| TLR4 | Asp299Gly (A/G) | WT | 2 (1,4%) | 4 (3,8%) | 0,028 |
| Hetero | 40 (28,4%) | 16 (15,4%) | |||
| Homo | 99 (70,2%) | 84 (80,8%) | |||
| Thr399Ile (T/C) | WT | 2 (1,4%) | 4 (3,8%) | 0,028 | |
| Hetero | 40 (28,4%) | 16 (15,4%) | |||
| Homo | 99 (70,2%) | 84 (80,8%) | |||
| TLR9 | T1237C | Hetero | 59 (41,8%) | 33 (31,7%) | 0,106 |
| Homo | 82 (58,2%) | 71 (68,3%) | |||
| T1486C | Hetero | 59 (41,8%) | 33 (31,7%) | 0,106 | |
| Homo | 82 (58,2%) | 71 (68,3%) | |||
Association of TLR2, TLR4 and TLR9 variants and clinical outcome
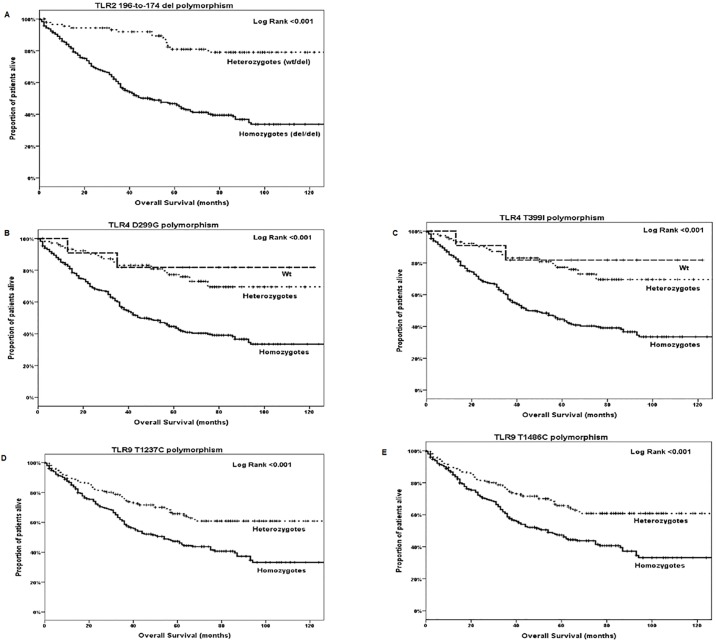
A relapse of the adjuvant treatment was observed in 64 (27.8%) of the enrolled adjuvant patients, whereas 197 (49.7%) of the stage IV patients, relapsed following front line treatment (Table 1). The median DFS and OS for stage IIA-IIIC patients was 18 months (95% CI: 12.4–23.7) and 155 months (95% CI: 75.0–235.0), respectively. There was no difference of DFS or OS according to the presence of different TLR genotypes. The median PFS and OS for stage IV patients was 8 months (95% CI: 7.1–8.9) and 31 months (95% CI: 25.1–36.9), respectively. There was again no difference of PFS or OS according to the presence of different TLR genotypes. However, the median OS for all patients was 75 months (95% CI: 56.8–93.1) and this prevailed a significant shorter OS in patients with TLR2 homozygous mutant (del/del) allele (p<0.001), TLR4 Asp299Gly (A/G) and Thr399Ile (T/C) homozygous mutant genotype (p<0.001 and p<0.001, respectively) or TLR9 T1237C and T1486C homozygous mutant genotype (p<0.001 and p<0.001, respectively) (Fig 2 and S3 Table).
Fig 2. Overall survival according to the detection of a) TLR2–196 to -174 del, b) TLR4 Asp299Gly (A/G), c) TLR4 Thr399Ile (T/C), d) TLR9 T1276C and e) TLR4 T1486C, respectively.
Discussion
The TLR pathway, through signaling by the myeloid differentiation primary response gene 88 (MyD88)-dependent pathway, increases the risk of colitis (or other inflammatory bowel diseases; IBDs)-associated CRC due to commensal gut microbiota [20–22]. In the present study, we aimed to evaluate the detection of TLR2 (-196 to -174 del), TLR4 (Asp299Gly and Thr399Ile) and TLR9 (T1237C and T1438C) polymorphisms in CRC patients.
The TLR2 gene is located on the chromosome 4 and the -196 to -174 del (a 22 bp nucleotide deletion) polymorphism alters the promoter activity of this gene. TLR2 del/del genotype is reported to show decreased transactivation of responsive promoters [11]. Previous studies investigated the likely association between this -196 to -174 del variant and nasopharyngeal cancer risk; the authors showed a significant association, concluding that the del/del genotype may represent a potential biomarker for nasopharyngeal cancer risk [23]. In another study conducted in Brazil, the authors showed that the frequency of heterozygous (ins/del) and homozygous mutant (del/del) alleles was significantly different between gastric cancer patients and healthy blood donors [24]; whereas others did not find any significant difference between gastric cancer patients and control groups for this variant [25–26]. An explanation to these contradictory results may be the enrolment of patients with distinct or ethnic differences among these studies. Nihon-Yanagi et al., suggest that TLR2 activation may play a role in sporadic CRC [9]. However, its role in CRC is also ambiguous. Studies have shown no significant differences in CRC development between wild type and TLR2 deficient mice [8, 27]; whereas, others have shown an increase in tumor growth and of IL-6, IL-17A and STAT3 levels in TLR2-deficient CRC mice, and a protective role of TLR against colitis-associated CRC [8, 28]. Our results are consistent with these studies since we observed higher frequencies of the TLR2 del allele in the CRC patients group compared with the control groups, highlighting the role of this polymorphism in colorectal carcinogenesis. Moreover, higher frequencies of the TLR2 del allele were detected in advanced CRC patients compared with the stage IIA-IIIC, emphasizing the role of this polymorphism in CRC progression and in patients’ overall survival.
Likewise, TLR4 has been known as indicative molecule for the detection of predisposition to cancer [9–10, 29–30]. The TLR4 gene is located in the chromosome 9 and single nucleotide polymorphisms in exon 3 lead to Asp299Gly and Thr399Ile substitutions [24]. The increased TLR4 levels have been correlated with poor prognosis in patients with gastric, prostate and colorectal cancer, although the results are still controversial. In fact, one study showed no association with the risk of gastric cancer [31], while another study observed that both polymorphisms were associated with gastric cancer development [32], despite both studies were performed in the same ethnic group. Moreover, studies have demonstrated that high expression of TLR4/MyD88 is associated with liver metastases and is an independent predictor for poor prognosis, both in colitis-associated and in sporadic CRC cases [33]. Controversial results have been shown by Davoodi and Seow, suggesting that both TLR4 Asp299Gly and Thr399Ile alleles are not associated with CRC risk [34]. Other studies showed that the detection TLR4 Asp299Gly polymorphism had a 4-fold higher risk for development of prostate cancer, among a North Indian population [35]. However, the results from a meta-analysis suggest that the TLR4 Asp299Gly polymorphism had a protective effect in prostate cancer; whereas, both TLR4 Asp299Gly and Thr399Ile polymorphisms were associated with an elevated gastrointestinal cancer risk [36–37]. In another study, the authors present a clear evidence for an association between TLR4 polymorphisms and CRC, suggesting that these polymorphisms could possibly serve as biomarkers for decision making in CRC treatment [38]. Our results are in accordance with other studies that associate the polymorphisms with patients’ outcome; in fact, our results suggest that the presence of both Asp299Gly and Thr399Ile polymorphisms were significantly associated with cancer development and progression, due to the higher frequency of these polymorphisms in CRC patients, and especially of those with metastatic disease. Moreover, the expression of either Asp299Gly or Thr399Ile homozygote mutant polymorphism had a great impact on the patients’ survival.
The TLR9 gene is located on chromosome 3. Several polymorphisms have been identified, which two of them are located in the promoter; T1237C in exon 2 and T1486C in intron 1 [39]. Several studies have shown that binding of TLR9 to CD4 cells can enhance their survival and therefore could potentiate antitumor responses by prolonging T-cell activity [40]. Moreover, it has been reported that the enhanced longevity of TLR9-stimulated mouse T cells in vitro was dependent on NF-κB activation and was associated with up-regulation of the antiapoptotic protein Bcl-xL[41]. Several TLR9 polymorphisms have been associated with susceptibility to inflammatory processes [42–43], thus highlighting the role of this gene in immune responses for gut homeostasis and the development of chronic inflammation. It has also been demonstrated that among others, TLR9 expression increases in active ulcerative colitis patients, and that the mRNA levels positively correlate with the severity of intestinal inflammation as well as with inflammatory cytokines [44–45]. Regarding the connection between polymorphisms in TLR9 genes and the risk of CRC, not much is known. It has been reported that oligodeoxynucleotides targeting TLR9 oppositely modulate DNA repair genes in tumor versus immune cells and enhance the antitumor activity of DNA-damaging chemotherapy and radiation therapy in preclinical mouse models [46]. Moreover, it has been demonstrated that TLR9 stimulates a series of immune responses, promotes angiogenesis and disease progression while decreasing patients’ survival [47–48]. In our study, both TLR9 T1237C and T1486C polymorphisms were more frequent in CRC patients compared to the control groups, and especially in stage IV patients, indicating their role in cancer development and progression.
A critical role in the resistance to treatment in patients with lung, pancreatic and colorectal cancer is played by continuous activation of the Ras/MAPK pathway because of mutations in codon 12 of the KRAS gene [49]. In our study, we also aimed to associate the frequency of TLR2, TLR4 and TLR9 polymorphisms in patients with different KRAS status. We found a significant association between both the TLR4 Asp299Gly and Thr399Ile polymorphisms with the KRAS mutant patients; however, no association was observed between any of the TLR2 or TLR9 polymorphisms and the KRAS status. It is to note that KRAS status was not determined for about 40% of the patients enrolled in the study, and these results have to be treated with caution.
In conclusion, our findings indicate a significant role of TLR2–196 to 174 del, TLR4 Asp299Gly, TLR4 Thr399Ile, TLR9 T1273C and TLR9 T1486C polymorphic variants with susceptibility to CRC development and disease progression, thus affecting patients’ survival. Due to the retrospective design, the lack of validation group and missing data regarding molecular subtypes of the enrolled tumors’ specimens, these results should be interpreted with caution and may serve mostly as hypothesis generated data for the rational design of future studies. Clearly, prospective studies are needed to better understand which microbial species are linked to cancer predisposition and which species are linked to positive outcomes. Moreover, further studies are necessary to the understanding of the way TLRs boost angiogenesis aiming the development of new anticancer agents and targeted treatments.
Supporting information
(DOCX)
(DOCX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was fully supported by research grants from the Hellenic Society of Medical Oncology (HeSMO; https://www.hesmo.gr/en/) and the Gastrointestinal Cancer Study Group (GIC-SG; https://www.emkapes.gr/?lang=en). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. Epub 2017/01/06. doi: 10.3322/caac.21387 . [DOI] [PubMed] [Google Scholar]
- 2.Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136(6):2015–31. Epub 2009/05/23. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tannock GW. Molecular analysis of the intestinal microflora in IBD. Mucosal Immunol. 2008;1 Suppl 1:S15–8. Epub 2008/12/23. doi: 10.1038/mi.2008.54 . [DOI] [PubMed] [Google Scholar]
- 4.Krishnan J, Selvarajoo K, Tsuchiya M, Lee G, Choi S. Toll-like receptor signal transduction. Exp Mol Med. 2007;39(4):421–38. Epub 2007/10/16. doi: 10.1038/emm.2007.47 . [DOI] [PubMed] [Google Scholar]
- 5.Pandey S, Agrawal DK. Immunobiology of Toll-like receptors: emerging trends. Immunol Cell Biol. 2006;84(4):333–41. Epub 2006/07/13. doi: 10.1111/j.1440-1711.2006.01444.x . [DOI] [PubMed] [Google Scholar]
- 6.Yu L, Chen S. Toll-like receptors expressed in tumor cells: targets for therapy. Cancer Immunol Immunother. 2008;57(9):1271–8. Epub 2008/02/08. doi: 10.1007/s00262-008-0459-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medvedev AE. Toll-like receptor polymorphisms, inflammatory and infectious diseases, allergies, and cancer. J Interferon Cytokine Res. 2013;33(9):467–84. Epub 2013/05/17. doi: 10.1089/jir.2012.0140 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pradere JP, Dapito DH, Schwabe RF. The Yin and Yang of Toll-like receptors in cancer. Oncogene. 2014;33(27):3485–95. Epub 2013/08/13. doi: 10.1038/onc.2013.302 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nihon-Yanagi Y, Terai K, Murano T, Matsumoto T, Okazumi S. Tissue expression of Toll-like receptors 2 and 4 in sporadic human colorectal cancer. Cancer Immunol Immunother. 2012;61(1):71–7. Epub 2011/08/17. doi: 10.1007/s00262-011-1085-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slattery ML, Herrick JS, Bondurant KL, Wolff RK. Toll-like receptor genes and their association with colon and rectal cancer development and prognosis. Int J Cancer. 2012;130(12):2974–80. Epub 2011/07/28. doi: 10.1002/ijc.26314 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noguchi E, Nishimura F, Fukai H, Kim J, Ichikawa K, Shibasaki M, et al. An association study of asthma and total serum immunoglobin E levels for Toll-like receptor polymorphisms in a Japanese population. Clin Exp Allergy. 2004;34(2):177–83. Epub 2004/02/28. . [DOI] [PubMed] [Google Scholar]
- 12.Gunter MJ, Stolzenberg-Solomon R, Cross AJ, Leitzmann MF, Weinstein S, Wood RJ, et al. A prospective study of serum C-reactive protein and colorectal cancer risk in men. Cancer Res. 2006;66(4):2483–7. Epub 2006/02/21. doi: 10.1158/0008-5472.CAN-05-3631 . [DOI] [PubMed] [Google Scholar]
- 13.Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25(2):187–91. Epub 2000/06/03. doi: 10.1038/76048 . [DOI] [PubMed] [Google Scholar]
- 14.Brand S, Staudinger T, Schnitzler F, Pfennig S, Hofbauer K, Dambacher J, et al. The role of Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms and CARD15/NOD2 mutations in the susceptibility and phenotype of Crohn’s disease. Inflamm Bowel Dis. 2005;11(7):645–52. Epub 2005/06/24. . [DOI] [PubMed] [Google Scholar]
- 15.Oostenbrug LE, Drenth JP, de Jong DJ, Nolte IM, Oosterom E, van Dullemen HM, et al. Association between Toll-like receptor 4 and inflammatory bowel disease. Inflamm Bowel Dis. 2005;11(6):567–75. Epub 2005/05/21. . [DOI] [PubMed] [Google Scholar]
- 16.Shen X, Shi R, Zhang H, Li K, Zhao Y, Zhang R. The Toll-like receptor 4 D299G and T399I polymorphisms are associated with Crohn’s disease and ulcerative colitis: a meta-analysis. Digestion. 2010;81(2):69–77. Epub 2010/01/23. doi: 10.1159/000260417 . [DOI] [PubMed] [Google Scholar]
- 17.Chuang TH, Ulevitch RJ. Cloning and characterization of a sub-family of human toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw. 2000;11(3):372–8. Epub 2000/10/07. . [PubMed] [Google Scholar]
- 18.Harsch M, Bendrat K, Hofmeier G, Branscheid D, Niendorf A. A new method for histological microdissection utilizing an ultrasonically oscillating needle: demonstrated by differential mRNA expression in human lung carcinoma tissue. Am J Pathol. 2001;158(6):1985–90. Epub 2001/06/08. doi: 10.1016/S0002-9440(10)64669-X . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahara T, Arisawa T, Wang F, Shibata T, Nakamura M, Sakata M, et al. Toll-like receptor 2–196 to 174del polymorphism influences the susceptibility of Japanese people to gastric cancer. Cancer Sci. 2007;98(11):1790–4. Epub 2007/08/23. doi: 10.1111/j.1349-7006.2007.00590.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niedzielska I, Niedzielski Z, Tkacz M, Orawczyk T, Ziaja K, Starzewski J, et al. Toll-like receptors and the tendency of normal mucous membrane to transform to polyp or colorectal cancer. J Physiol Pharmacol. 2009;60 Suppl 1:65–71. Epub 2009/07/23. . [PubMed] [Google Scholar]
- 21.Kinugasa T, Akagi Y. Status of colitis-associated cancer in ulcerative colitis. World J Gastrointest Oncol. 2016;8(4):351–7. Epub 2016/04/21. doi: 10.4251/wjgo.v8.i4.351 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen R, Alvero AB, Silasi DA, Steffensen KD, Mor G. Cancers take their Toll—the function and regulation of Toll-like receptors in cancer cells. Oncogene. 2008;27(2):225–33. Epub 2008/01/08. doi: 10.1038/sj.onc.1210907 . [DOI] [PubMed] [Google Scholar]
- 23.Makni L, Messadi A, Zidi S, Gazouani E, Mezlini A, Yacouni-Loueslati B. TLR2 (-196 to -174 Ins/Del) and TLR3 (1377C>T) as biomarkers for nasopharyngeal cancer in Tunisia. Turkish Journal of Medical Sciences. 2017:1216–22. Epub 1/04/2017. doi: 10.3906/sag-1608-17 [DOI] [PubMed] [Google Scholar]
- 24.de Oliveira JG, Silva AE. Polymorphisms of the TLR2 and TLR4 genes are associated with risk of gastric cancer in a Brazilian population. World J Gastroenterol. 2012;18(11):1235–42. Epub 2012/04/03. doi: 10.3748/wjg.v18.i11.1235 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hishida A, Matsuo K, Goto Y, Naito M, Wakai K, Tajima K, et al. No associations of Toll-like receptor 2 (TLR2) -196 to -174del polymorphism with the risk of Helicobacter pylori seropositivity, gastric atrophy, and gastric cancer in Japanese. Gastric Cancer. 2010;13(4):251–7. Epub 2010/12/04. doi: 10.1007/s10120-010-0567-y . [DOI] [PubMed] [Google Scholar]
- 26.Liu S, Wang X, Shi Y, Han L, Zhao Z, Zhao C, et al. Toll-like receptor gene polymorphisms and susceptibility to Epstein-Barr virus-associated and -negative gastric carcinoma in Northern China. Saudi J Gastroenterol. 2015;21(2):95–103. Epub 2015/04/07. doi: 10.4103/1319-3767.153832 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salcedo R, Worschech A, Cardone M, Jones Y, Gyulai Z, Dai RM, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J Exp Med. 2010;207(8):1625–36. Epub 2010/07/14. doi: 10.1084/jem.20100199 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe EL, Crother TR, Rabizadeh S, Hu B, Wang H, Chen S, et al. Toll-like receptor 2 signaling protects mice from tumor development in a mouse model of colitis-induced cancer. PLoS One. 2010;5(9):e13027 Epub 2010/10/05. doi: 10.1371/journal.pone.0013027 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz E, Frees KL, Schwartz DA. Determination of the TLR4 genotype using allele-specific PCR. Biotechniques. 2001;31(1):22–4. Epub 2001/07/24. . [DOI] [PubMed] [Google Scholar]
- 30.Gatti G, Quintar AA, Andreani V, Nicola JP, Maldonado CA, Masini-Repiso AM, et al. Expression of Toll-like receptor 4 in the prostate gland and its association with the severity of prostate cancer. Prostate. 2009;69(13):1387–97. Epub 2009/06/06. doi: 10.1002/pros.20984 . [DOI] [PubMed] [Google Scholar]
- 31.Garza-Gonzalez E, Bosques-Padilla FJ, Mendoza-Ibarra SI, Flores-Gutierrez JP, Maldonado-Garza HJ, Perez-Perez GI. Assessment of the toll-like receptor 4 Asp299Gly, Thr399Ile and interleukin-8–251 polymorphisms in the risk for the development of distal gastric cancer. BMC Cancer. 2007;7:70 Epub 2007/04/28. doi: 10.1186/1471-2407-7-70 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trejo-de la OA, Torres J, Perez-Rodriguez M, Camorlinga-Ponce M, Luna LF, Abdo-Francis JM, et al. TLR4 single-nucleotide polymorphisms alter mucosal cytokine and chemokine patterns in Mexican patients with Helicobacter pylori-associated gastroduodenal diseases. Clin Immunol. 2008;129(2):333–40. Epub 2008/08/30. doi: 10.1016/j.clim.2008.07.009 . [DOI] [PubMed] [Google Scholar]
- 33.Wang EL, Qian ZR, Nakasono M, Tanahashi T, Yoshimoto K, Bando Y, et al. High expression of Toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. Br J Cancer. 2010;102(5):908–15. Epub 2010/02/11. doi: 10.1038/sj.bjc.6605558 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davoodi H, Seow HF. Variant Toll-like receptor4 (Asp299Gly and Thr399Ile alleles) and Toll-like receptor2 (Arg753Gln and Arg677Trp alleles) in colorectal cancer. Iran J Allergy Asthma Immunol. 2011;10(2):91–9. Epub 2011/06/01. . [PubMed] [Google Scholar]
- 35.Priyadarshini A, Chakraborti A, Mandal AK, Singh SK. Asp299Gly and Thr399Ile polymorphism of TLR-4 gene in patients with prostate cancer from North India. Indian J Urol. 2013;29(1):37–41. Epub 2013/05/15. doi: 10.4103/0970-1591.109982 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jing JJ, Li M, Yuan Y. Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms in cancer: a meta-analysis. Gene. 2012;499(2):237–42. Epub 2012/03/24. doi: 10.1016/j.gene.2012.03.045 . [DOI] [PubMed] [Google Scholar]
- 37.Sheng WY, Yong Z, Yun Z, Hong H, Hai LL. Toll-like receptor 4 gene polymorphisms and susceptibility to colorectal cancer: a meta-analysis and review. Arch Med Sci. 2015;11(4):699–707. Epub 2015/09/01. doi: 10.5114/aoms.2015.53288 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semlali A, Reddy Parine N, Arafah M, Mansour L, Azzi A, Al Shahrani O, et al. Expression and Polymorphism of Toll-Like Receptor 4 and Effect on NF-kappaB Mediated Inflammation in Colon Cancer Patients. PLoS One. 2016;11(1):e0146333 Epub 2016/01/16. doi: 10.1371/journal.pone.0146333 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng MW, Lau CS, Chan TM, Wong WH, Lau YL. Polymorphisms of the toll-like receptor 9 (TLR9) gene with systemic lupus erythematosus in Chinese. Rheumatology (Oxford). 2005;44(11):1456–7. Epub 2005/09/29. doi: 10.1093/rheumatology/kei120 . [DOI] [PubMed] [Google Scholar]
- 40.Rahman AH, Taylor DK, Turka LA. The contribution of direct TLR signaling to T cell responses. Immunol Res. 2009;45(1):25–36. Epub 2009/07/15. doi: 10.1007/s12026-009-8113-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gelman AE, Zhang J, Choi Y, Turka LA. Toll-like receptor ligands directly promote activated CD4+ T cell survival. J Immunol. 2004;172(10):6065–73. Epub 2004/05/07. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lammers KM, Ouburg S, Morre SA, Crusius JB, Gionchett P, Rizzello F, et al. Combined carriership of TLR9-1237C and CD14-260T alleles enhances the risk of developing chronic relapsing pouchitis. World J Gastroenterol. 2005;11(46):7323–9. Epub 2006/01/27. doi: 10.3748/wjg.v11.i46.7323 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torok HP, Glas J, Endres I, Tonenchi L, Teshome MY, Wetzke M, et al. Epistasis between Toll-like receptor-9 polymorphisms and variants in NOD2 and IL23R modulates susceptibility to Crohn’s disease. Am J Gastroenterol. 2009;104(7):1723–33. Epub 2009/05/21. doi: 10.1038/ajg.2009.184 . [DOI] [PubMed] [Google Scholar]
- 44.Sanchez-Munoz F, Fonseca-Camarillo G, Villeda-Ramirez MA, Miranda-Perez E, Mendivil EJ, Barreto-Zuniga R, et al. Transcript levels of Toll-Like Receptors 5, 8 and 9 correlate with inflammatory activity in Ulcerative Colitis. BMC Gastroenterol. 2011;11:138 Epub 2011/12/22. doi: 10.1186/1471-230X-11-138 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan Y, Liu B. Expression of Toll-like receptors in the mucosa of patients with ulcerative colitis. Exp Ther Med. 2015;9(4):1455–9. Epub 2015/03/18. doi: 10.3892/etm.2015.2258 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sommariva M, De Cecco L, De Cesare M, Sfondrini L, Menard S, Melani C, et al. TLR9 agonists oppositely modulate DNA repair genes in tumor versus immune cells and enhance chemotherapy effects. Cancer Res. 2011;71(20):6382–90. Epub 2011/09/01. doi: 10.1158/0008-5472.CAN-11-1285 . [DOI] [PubMed] [Google Scholar]
- 47.Belmont L, Rabbe N, Antoine M, Cathelin D, Guignabert C, Kurie J, et al. Expression of TLR9 in tumor-infiltrating mononuclear cells enhances angiogenesis and is associated with a worse survival in lung cancer. Int J Cancer. 2014;134(4):765–77. doi: 10.1002/ijc.28413 Epub 2013 Sep 4. [DOI] [PubMed] [Google Scholar]
- 48.Holldack J. Toll-like receptors as therapeutic targets for cancer. Drug Discov Today. 2014;19(4):379–82. doi: 10.1016/j.drudis.2013.08.020 Epub Sep 3. [DOI] [PubMed] [Google Scholar]
- 49.Rosa R, Melisi D, Damiano V, Bianco R, Garofalo S, Gelardi T, et al. Toll-like receptor 9 agonist IMO cooperates with cetuximab in K-ras mutant colorectal and pancreatic cancers. Clin Cancer Res. 2011;17(20):6531–41. Epub 2011/09/06. doi: 10.1158/1078-0432.CCR-10-3376 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.