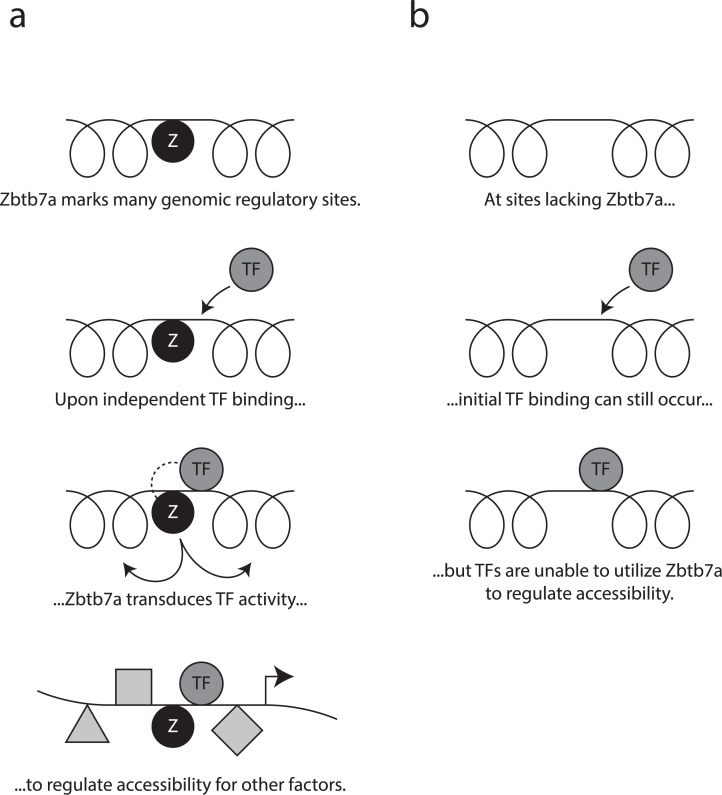
Fig 7. Scheme of Zbtb7a function at gene-regulatory elements.
Cartoon outlining role of Zbtb7a described in this paper. Principal experimental evidence for each depicted step is shown in the indicated figures. (A) Top row: Zbtb7a binds to many genomic regulatory sites, including promoters and enhancers (see Fig 2C and 2D). Zbtb7a binding is independent of the presence of client TFs, and binding may occur before client TF recruitment (see Fig 6A). Second row: Zbtb7a-utilising client TFs bind independently to neighbouring genomic sites. This may occur under normal, steady-state conditions or in response to stimulation (see Figs 4A and 6B). Third row: Zbtb7a transduces TF-dependent changes in local chromatin accessibility (see Figs 4C, 5D, 6C, 6D, 6E and 6F). In the case of p65, this is triggered by the interaction between Zbtb7a and the TA3 region of p65 (see Fig 5F). Bottom row: Zbtb7a-transduced accessibility allows binding of secondary TFs and contributes to normal gene activation (see Figs 4D, 6G and S3L and S3M Fig). (B) At genomic sites that lack Zbtb7a binding (which could occur due to the natural genomic distribution of Zbtb7 or through experimental manipulation), client TF binding is unimpaired (see Fig 6B), but Zbtb7a-dependent regulation of accessibility is abolished (see Figs 4C, 6C, 6D, 6E and 6F). TF, transcription factor.