Figure 5.
High GATA6 Expression Results in Endoderm Differentiation Bias at Population and Single-Cell Level
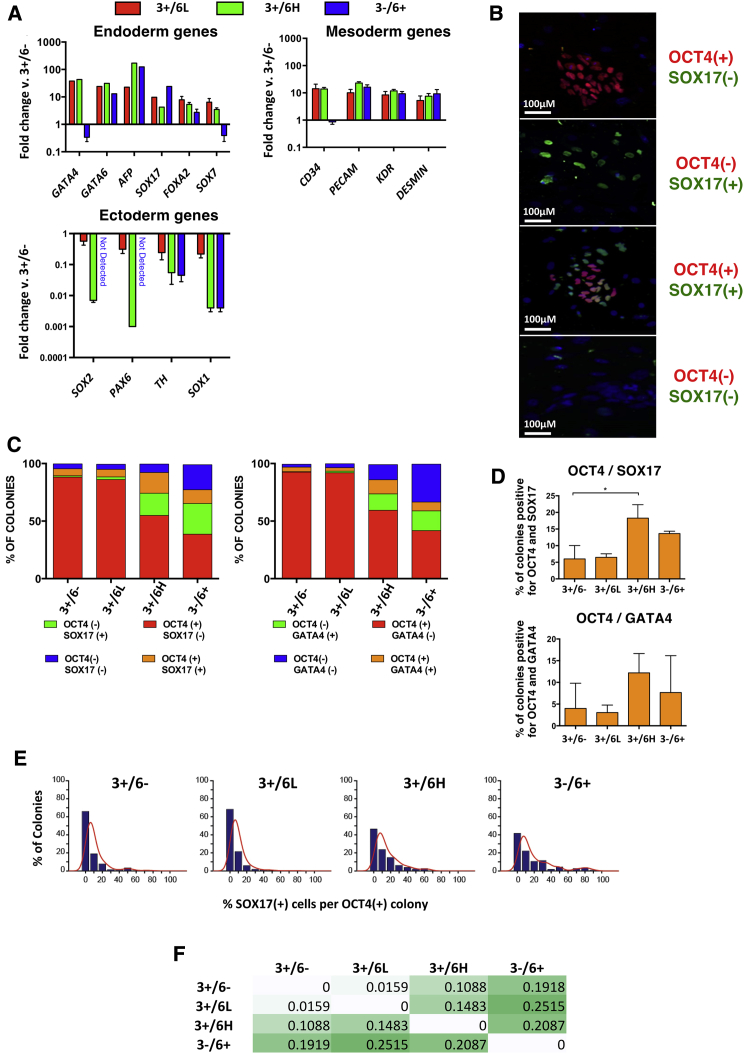
(A) qPCR of differentiating cells from the 3+/6L (red), 3+/6H (green), 3-/6+ (blue) fractions in a non-directed EB differentiation assay, shown as fold change against differentiating cells from 3+/6− fraction, for genes expressed in endoderm (top left), mesoderm (top right), and ectoderm (bottom left). Beta-actin was the normalizing gene. Error bars represent three biological replicates.
(B) Representative images of colonies derived from 3+/6−, 3+/6L, 3+/6H, and 3−/6+ fractions. Images were taken at ×10 magnification on an InCell Analyzer 2000 and automated quantitative analysis performed using developer toolbox software. The same algorithms were used for each technical and biological repeat and the process was automated to eliminate human bias.
(C) Quantification of colony types from 3+/6−, 3+/6L, 3+/6H, and 3−/6+ fractions showing the percentage of colonies per fraction with colony phenotype shown in (B) from three biological repeats.
(D) Percentage of colonies containing OCT4 and SOX17 (top graph) or OCT4 and GATA4 (bottom graph) positive cells only. Significance was calculated using t test of three biological replicates and stars represent degree of significance (∗p < 0.05). Numbers for each fraction: 3+/6− = 83, 3+/6L = 103, 3+/6H = 122, 3−/6+ = 66.
(E) Histogram showing the distribution of SOX17(+) cells in OCT4-positive colonies resulting from single cells from 3+/6−, 3+/6L, 3+/6H, and 3−/6+ fractions. Positive colonies include at least two OCT4(+) cells. Counts are shown as a bar plot (blue) with superimposed estimated nonparametric distribution (red).
(F) Kullback-Leibler symmetric divergence between SOX17-associated distributions in OCT4-positive colonies. This measure increases with reduced similarity between distributions; zero indicates identical distributions.