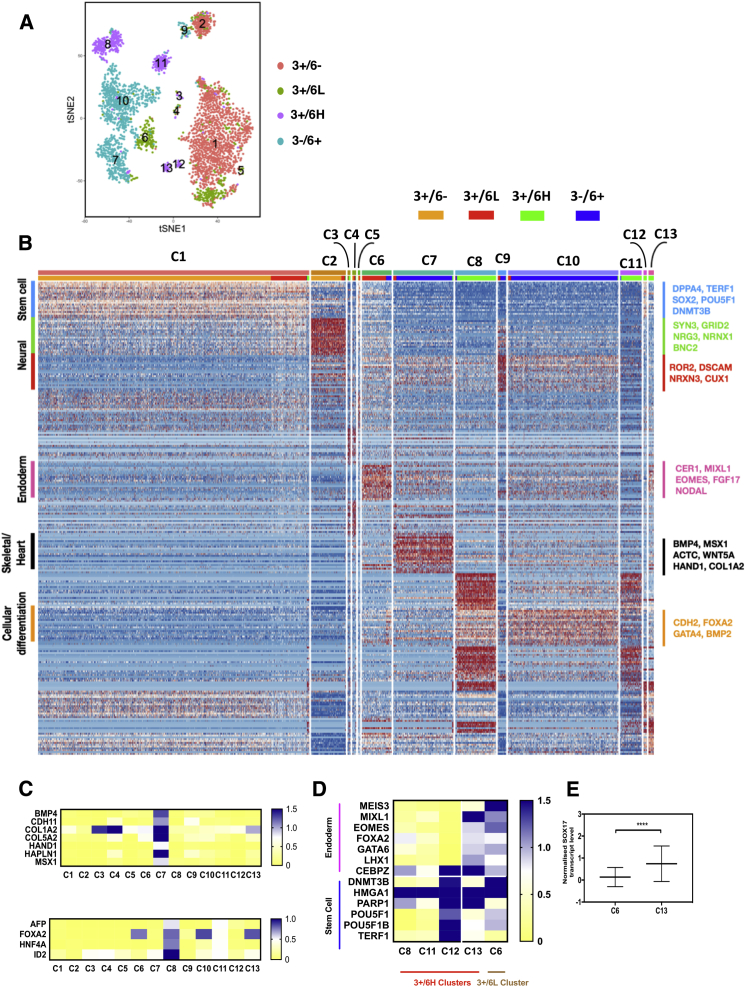
Figure 6.
Single-Cell Transcriptomic Analysis of Endodermally Biased hESCs
(A) tSNE analysis of the four cellular subsets representing 13 putative clusters separated according to gene expression per single cells. Single cells are represented by individual dots and are colored according to cell fraction library. Numbers represent cluster number assigned arbitrarily.
(B) Heatmap of the top 30 most differentially expressed genes between the 13 individual putative clusters, as described in Figure 6A. Color scheme is based on Z score distribution from −2 (blue) to +2 (red). Right margin color bars represent gene sets specific to each cluster. Left margin color bars represent top Gene Ontology terms of the top 30 most differentially expressed genes for each cluster.
(C) Heatmap of the average expression of genes typically associated with later developmental processes including heart/skeletal (top) and hepatic (lower) lineages across the 13 clusters. Color scheme is based on the averaged normalized expression of each gene from no expression (yellow) to expression (blue).
(D) Heatmap of the average expression of genes associated with both the stem cells and early differentiating cells across all the individual 3+/6H clusters and the 3+/6L cluster. Color scheme is based on the averaged normalized expression of each gene from no expression (yellow) to expression (blue).
(E) Scatter dot plot to show the mean, upper, and lower limit expression of SOX17 between cluster 6 of the 3+/6L and cluster 13 of the 3+/6H fractions. Student's t test was used to determine statistical significance of ∗∗∗∗p > 0.0001.