Figure 1.
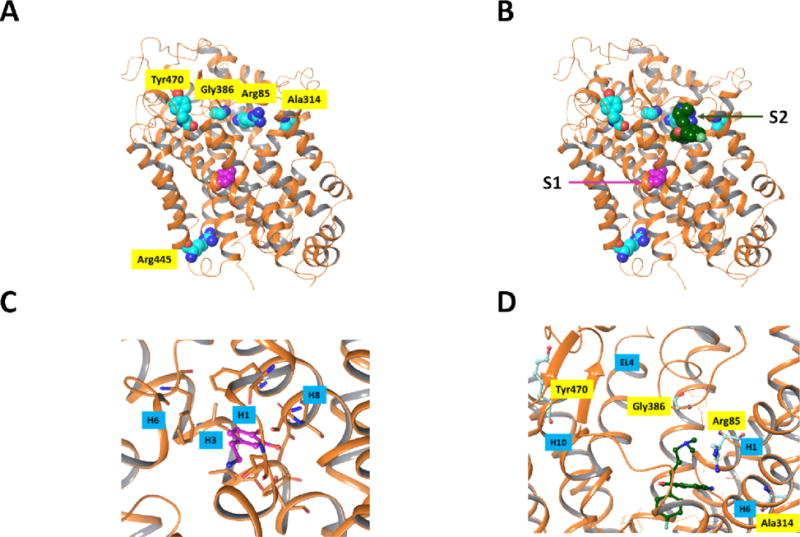
Positions of residues mutated in previous DTDS studies. The protein is shown in ribbon rendering and atoms which are displayed are rendered as space filling structures. Protein carbon atoms are aqua colored and dopamine carbon atoms are depicted in magenta. The X-ray structure is that of dDAT20 [PDB entry 4XP1] and the residues which were mutated are indicated using the hDAT numbering. A) All mutated residues, with the exception of R445, are in the upper region of the protein. None of the mutated residues are in close contact with the dopamine substrate in the primary binding site. B) Based on a superposition of dDAT and the recently published X-ray structure of SERT17 with SCIT occupying both the primary binding site (S1) as well as the vestibule binding site on the intracellular entryway to SERT (S2) it can be seen that the 4 mutated residues in the upper portion of the protein are in the vestibule region and could potentially interfere with the substrate binding at this stopover site. Only the SCIT ligand of the hSERT X-ray structure is shown. C) Closeup view of the primary substrate binding site in dDAT showing the residues close to the DA substrate in stick models. Nearby helices H1, H3, H6 and H8 are indicated. D) Closeup view of the “vestibule” region in dDAT in A). For reference, the residues of dDAT corresponding to the dDAT mutants observed in DTDS are shown as aqua ball and stick structures. Nearby helices H1, H6 and H10 and extracellular loop EL4 are indicated. Molecular graphics were generated with Maestro21.
