Figure 5.
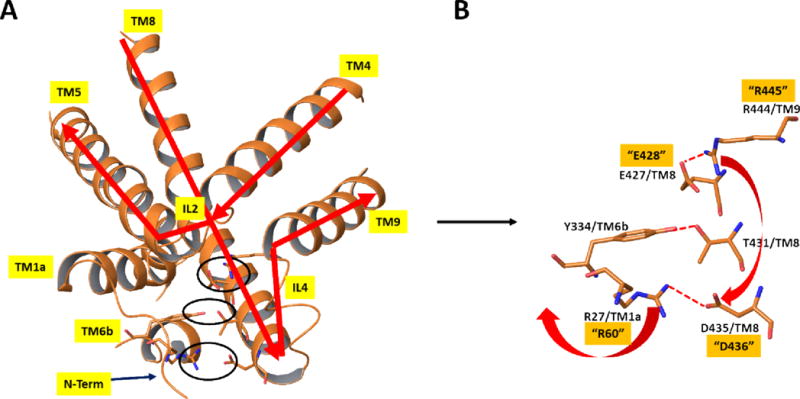
X-ray structure of dDAT19 [PDB entry 4XP1] in the same orientation and from the same view as in Figure 4. A) Close-up view of the protein in the region discussed using ribbon representation of the protein and depicting key polar interactions in stick figures. B) Stick figure representation of key polar interactions in the extracellular side responsible for maintaining the closed-inward (open-outward) protein form in the region shown. Numbering of residues is based on dDAT, as in the X-ray structure. The corresponding hDAT numbering for the residues mutated in the current study are depicted with quotes. Red arrows indicate the proposed structural rearrangements which would occur if the salt bridge between E427 and R444 were broken. Specifically, R444 could alternatively form a salt bridge with D435, thereby disrupting the salt bridge between D435 and R27 and, in turn releasing the N-terminus to form the inward open structure. Molecular graphics were generated with Maestro21.
