Figure 1. Introduction of F2483I mutation in RYR2 gene of hiPSC.
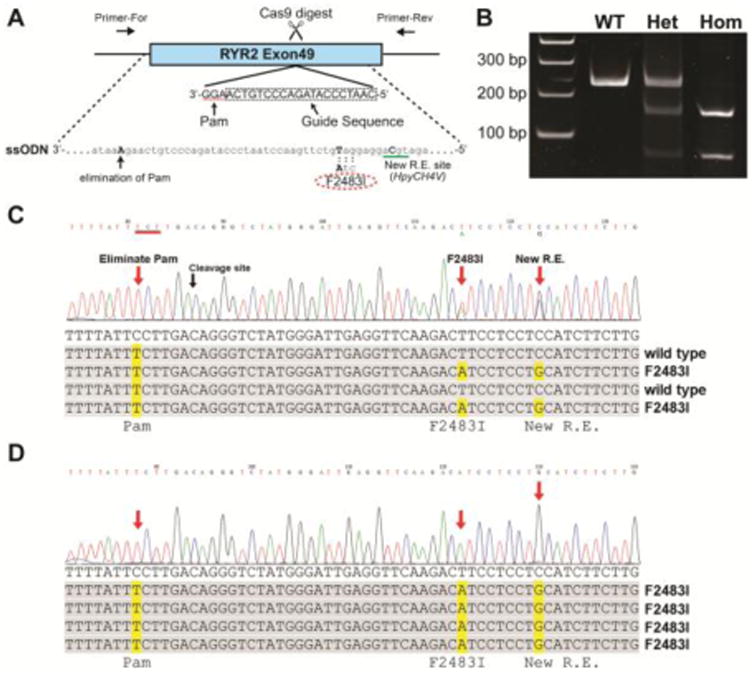
(A) Schematic of gene-editing. Exon49 of RYR2 gene in wild type hiPSC was targeted by CRISPR/Cas9 technique. Guide sequence shown in dotted box was cloned into pX459 vector to express sgRNA guiding Cas9 exonuclease to the targeted Pam sequence (highlighted by dotted line). The ssODN was used to introduce mutations during homology-directed repair of the Cas9-digested genome. The ssODN carried F2483I mutation (TTC to ATC), a mutation creating HpyCH4V restriction enzyme site, and a mutation eliminating Pam sequence (AGG to AGA). Note that Guide sequence and Pam sequence were on the anti-strand of the genome, thus ssODNA was also anti-strand sequence. (B) PCR analysis of the targeted hiPSCs. The gene-edited regions were amplified from genomic DNA by PCR, followed by restriction digest by HpyCH4V. Full size of PCR product was 228 bp (wild type). PCR products from the heterozygous (Het) mutant were partially digested by the enzyme, while ones from homozygous (Hom) mutant were completely digested in to two fragments, 150 and 78 bp. (C) Sequencing of PCR products from the heterozygous mutant. Direct sequencing of the whole PCR products showed double peaks at F2483I and new restriction enzyme (R.E.) sites, indicating that only one allele carries these mutations. Single peak on the Pam sequence mutation site, suggesting that both gene allele has this mutation. Sequencing of cloned PCR products confirmed these mutations (lower panel). (D) Sequencing of PCR products from the homozygous mutant. All three mutation sites showed single peaks in whole PCR sequencing, indicating both allele carries these three mutations. Sequencing of cloned PCR products confirmed this.
