Figure 2.
Optopatch Measurement and Analysis Pipeline
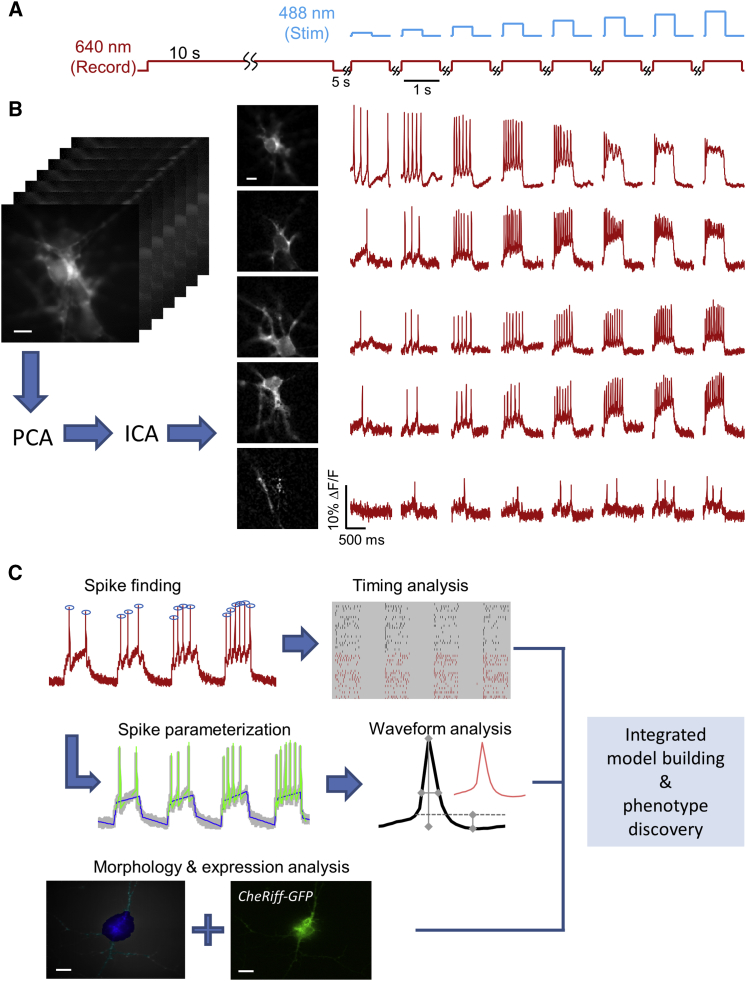
(A) Cells were subject to 10 s of unstimulated recording to measure spontaneous activity (red), and then to eight stimulation pulses of 500 ms duration and increasing intensity (blue).
(B) Activity-based movie segmentation. Image stacks were filtered spatially and temporally, then processed via principal components analysis (PCA) followed by independent components analysis (ICA) to identify clusters of pixels whose fluorescence values co-varied in synchrony. The movie was decomposed into a sum of overlapping neuron images, each with its own spiking pattern.
(C) Parameterization pipeline. Spikes were identified in the fluorescence traces. Spiking patterns were analyzed within stimuli, between stimuli, and between populations. AP waveforms were also parameterized, enabling comparison of width, height, and after-polarization within and between cells. The results of the segmentation in the spatial domain enabled measurement of morphological features (soma versus dendrite) and of CheRiff-EGFP expression level. Finally, all of this information was integrated to build a coherent picture of phenotypic differences between mutant and control cell lines.
All scale bars, 10 μm. See also Figures S2–S4.