Dear Editor,
Loss or dysfunction of cardiomyocytes lead to heart failure which is a leading cause of death worldwide.1 It is widely known that the hearts of adult mammals have a very limited regeneration capacity.2 A number of cell transplantation strategies have been proposed to restore cardiac functions, including the transplantation of primary cardiac progenitor cells (CPCs), or pluripotent stem cell-derived CPC-like, or cardiomyocyte-like cells.2 However, the former approach suffers from the limited availability of cell sources and the latter has yet to overcome the differentiation efficiency, immune compatibility and possible tumorigenesis problems.3 Functional cardiomyocytes could also be generated by direct reprogramming of non-muscle cells with forced expression of cardiac transcription factors or microRNAs.4 More interestingly, cardiomyocytes could be induced from non-myocytes in the heart in vivo by local delivery of transcription factors Gata4, Mef2c, and Tbx5 after coronary ligation.5 However, viral vector-carried transcription factors are still not favorable in therapeutic applications.6
Small-molecule compounds provide a more clinically amendable way to induce somatic cell reprogramming. Several types of cells, including induced pluripotent stem cells, neural progenitor cells, neurons and endodermal progenitors, have been generated from various types of somatic cells with chemical cocktails.7 Recently we reported the generation of spontaneously beating cardiomyocyte-like cells (CiCMs) from mouse fibroblasts using only chemical cocktails.8 Shortly after that, human CiCMs were generated successfully from fibroblasts.9 However, it remains unclear whether these chemicals could induce in vivo cardiac reprogramming. Here we report that a chemical combination of CRFVPTM (C, CHIR99021; R, RepSox; F, Forskolin; V, VPA; P, Parnate; T, TTNPB; M, Rolipram) is able to induce the generation of cardiomyocytes from cardiac fibroblasts in vivo in normal adult mice. And this combination can also reduce the formation of fibrotic tissues after myocardial infarction (MI).
Our previous study demonstrated that CRFVPTM is very efficient in inducing the cardiac transdifferentiation in vitro.8 A series of drug delivery schemes (Schemes 1-5, Supplementary information, Figure S1a) was designed to test their efficacy in inducing cardiac reprogramming in vivo. Fsp1-Cre:R26RtdTomato mice were used to confirm the non-cardiomyocyte origin of the possibly emerging CiCMs.4, 5 Apart from labeling fibroblasts, Fsp1-driven transgene has been reported to label a number of other types of cells (including endothelial cells, vascular smooth muscle cells and hematopoietic cells) in the heart except cardiomyocytes.10 Immunofluorescence staining (Supplementary information, Figure S2) revealed that a small portion of the tdTomato+ cells express α-SMA (vascular smooth muscle marker10), vimentin (endothelial marker11), or CD11b (hematopoietic marker10), but none of the tdTomato+ cells express cTnI and Mef2C (cardiomyocyte markers), very similar to previous findings.10 Since the majority of tdTomato+ cells are fibroblasts, we still refer to these cells as cardiac fibroblasts for the sake of convenience. According to previous animal studies, CRFTM were given orally, and VP were given via intraperitoneal injection. Drug delivery with short intervals (once a day for 7 days, Scheme 1 and the end of Scheme 4) induced significant weight loss in mice, indicating a possible general toxic effect (Supplementary information, Figure S1b, e). Drug delivery with longer intervals, twice a week for two weeks (Scheme 2) or once a week for 4–6 weeks (Schemes 3 and 5), did not cause significant weight loss (Supplementary information, Figure S1c, d and f). Thus, most of the animals were treated as Scheme 5 (Supplementary information, Figure S1a), and they received CRFVPTM once a week for 6 weeks.
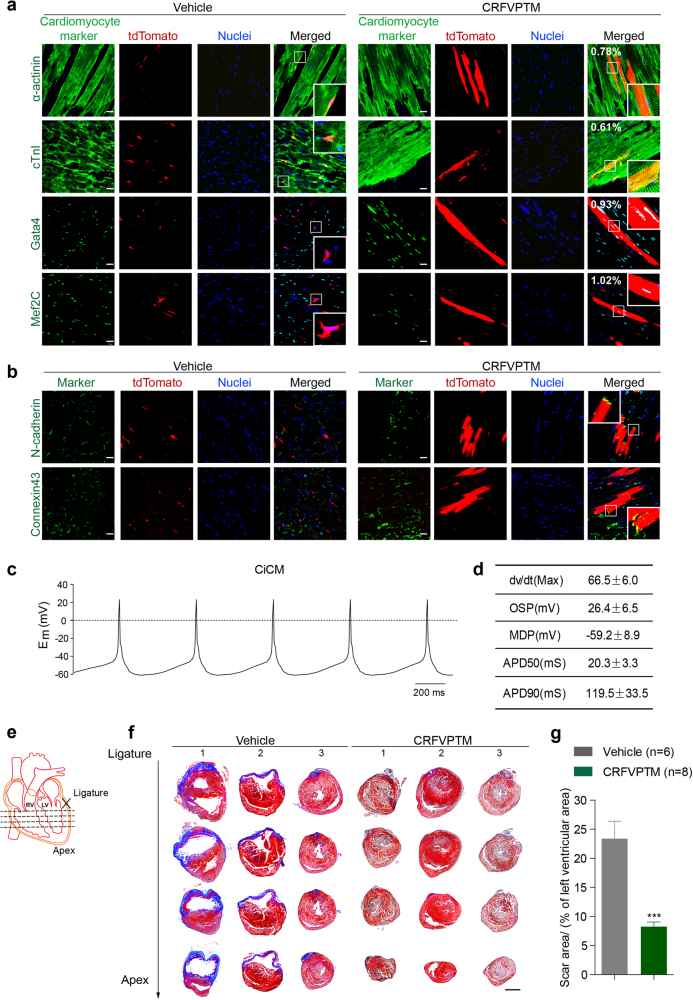
The animals were killed 6 weeks later, and the hearts were examined for possible cardiac reprogramming. In vehicle-treated mice, all the tdTomato+ cells were small, containing processes, resided next to large cardiomyocyte fibers and did not express any of the tested cardiomyocyte markers (Fig. 1a, left). We found that large, cardiomyocyte-like tdTomato+ cells appeared in CRFVPTM-given group (Fig. 1a, right). These long and rod-shaped tdTomato+ cells also expressed cardiomyocyte-specific markers, including α-actinin, cTnI, Gata4, and Mef2c, with well-formed sarcomeric structure very similar to the tdTomato− cardiomyocytes next to them (Fig. 1a, right). Approximately 0.78% of the tdTomato+ cells were α-actinin+ and large in size, and 1.02% of the tdTomato+ cells were Mef2C+. Hearts from these mice (Scheme 5) were also digested and the cells were cultured in single layer. No co-localization of tdTomato and cardiac markers could be observed in cells isolated from the hearts of vehicle-treated mice (Supplementary information, Figure S3). In contrast, 0.6–0.9% of the tdTomato+ cells from the hearts of the CRFVPTM-treated mice expressed cardiac markers (Supplementary information, Figure S3). Clear sarcomeric structure could be observed in the tdTomato+CiCMs (Supplementary information, Figure S3).
Fig. 1.
Direct reprogramming of adult cardiac fibroblasts into cardiomyocytes in vivo by chemical cocktails. a Eight-week-old Fsp1-cre:R26RtdTomato mice were given CRFVPTM or vehicle once a week for 6 weeks (Supplementary information, Figure S1a, Scheme 5). The animals were then killed and the cryosections of the hearts were stained with antibodies against Mef2C, Gata4, cTnI, and α-actinin. The numbers in the merged images of the drug-treated group represent the percent of tdTomato+ cells expressing various cardiac markers (five sections from each mouse were analyzed, n = 6 for control group and n = 8 for drug-treated group). Nuclei were stained with hoechst. Scale bar, 20 μm. b Immunofluorescence staining of N-cadherin and Connexin 43 in the cryosections of the hearts from Fsp1-cre:R26RtdTomato mice treated with CRFVPTM or vehicle once a week for 6 weeks. Nuclei were stained with hoechst. Scale bar, 20 μm. c Representative action potentials (APs) of the tdTomato+ CiCMs isolated from the hearts of mice treated with CRFVPTM once a week for 6 weeks. APs were recorded in current clamp mode at zero applied current. Em, membrane potential in millivolts. d AP parameters of the tdTomato+ CiCMs, including maximum upstroke velocity (dv/dt Max), over shoot potential (OSP), minimum diastolic potential (MDP), AP durations (APDs) at the level of 50% (APD50) and 90% repolarization (APD90). Data are means ± SEM (n = 7). e Schematic drawing of the positions of the four section levels related to the LAD ligation site. f Representative images (mice 1–3 in both groups) of Masson’s trichrome staining of the heart sections (as presented in e) from mice with LAD ligation and then treated with CRFVPTM or vehicle once a week for 6 weeks (blue areas represent fibrosis, and red areas represent normal tissue). Scale bar, 2 mm. g Quantification of fibrosis area (blue) relative to left ventricular area in heart sections with Masson’s trichrome staining. Four levels from each heart (e) and four slides from each level were measured (a total of 16 sections from each heart. n = 6 for vehicle group and n = 8 for drug-treated group). Data are presented as means ± SEM, ***P < 0.001
Similar results were also obtained in mice treated as Schemes 3 and 4 (Supplementary information, Figure S4), with slight difference in reprogramming efficiency. Furthermore, we found that in Schemes 1 and 2, tdTomato+ cells expressing cardiac-specific markers also emerged, but with a much lower efficiency (Supplementary information, Figure S5). Although these cells were much larger in size than the tdTomato+ fibroblasts, they were shorter and smaller than the endogenous tdTomato− adult cardiomyocytes (Supplementary information, Figure S5), indicating that the newly generated CiCMs were not mature and were still in transition state. In all Schemes, tdTomato+ cells expressing cardiomyocyte markers were not observed in vehicle controls (Fig. 1a; Supplementary information, Figures S4 and S5).
To exclude the possibility that the chemicals cause fibrosis and activate Fsp1-driven tdTomato expression in cardiomyocytes, which results in co-localization of tdTomato and cardiac markers, another transgenic mouse line, the α-MHC-cre:R26RtdTomato, in which the cardiomyocytes are labeled with tdTomato, were treated with CRFVPTM (Fig. S1a Scheme 5). No tdTomato+ cardiomyocytes were found to express Fsp1 after drug treatment (Supplementary information, Figure S6). In contrast, if the mice were subjected to MI, many tdTomato+ cardiomyocytes were found to be co-labeled with Fsp1 (Supplementary information, Figure S6). These data indicate that drug treatment do not cause fibrosis and activation of Fsp1 gene in cardiomyocytes in mice.
Further characterization of the CiCMs were conducted out in mice from Scheme 5. Connexin 43, the major gap junction protein in the heart, which is important for electrical coupling and synchronized contraction of the cardiomyocytes, was also expressed in the tdTomato+ CiCMs, suggesting good coupling of these cells with neighboring cardiomyocytes (Fig. 1b). N-cadherin, a cell surface Ca2+-dependent protein in intercalated disks, was also expressed at the cell border as an indicator of good coupling between CiCMs and the endogenous cardiomyocytes (Fig. 1b). Cardiomyocytes were isolated from these drug-treated mice and electrophysiological recordings were performed. The tdTomato+ CiCMs that we recorded all generated action potentials (APs) very similar to atrial cardiomyocytes (Fig. 1c, d).
Next we examined whether the chemical cocktail could induce cardiac reprogramming in skeletal muscle. The skeletal muscle cells expressed α-MHC and α-actinin (markers for both skeletal muscle and cardiomyocytes), and Nebulin (specific for skeletal muscle), but not Gata4 (specific for cardiomyocytes) (Supplementary information, Figure S7a). tdTomato+ fibroblasts could also be found in skeletal muscles (Supplementary information, Figure S7a). However, 6-week drug treatment did not turn activate expression of any muscle markers in the tdTomato+ cells (Supplementary information, Figure S7a). The tail tip, lung and liver were also examined. All these tissue or organs contained plenty of tdTomato+ cells, but none of the tdTomato+ cells expressed cardiomyocyte markers after drug treatment (Supplementary information, Figure S7b–d). Taken together, these data indicate that the native cardiac environment has a critical role in chemical-mediated cardiac reprogramming.
The formation of fibrotic tissue after MI causes irreversible damage to the heart function. Converting cardiac fibroblasts into cardiomyocytes not only enhances the regeneration of cardiomyocytes, but also reduces the formation of fibrotic tissues. To test this, cardiac injury was induced by coronary artery ligation in adult C57BL/6 mice (Fig. 1e). One week after MI, the mice were treated with CRFVPTM once a week for 6 weeks (Supplementary information, Figure S8a). Drug treatment did not cause noticeable weight change in the animals (Supplementary information, Figure S8b). MI typically causes cardiac hypertrophy and increases the heart/body weight ratio, but in CRFVPTM-treated MI mice, the heart/body weight ratio remained similar to the mock-operated group (Supplementary information, Figure S8c). Histological examination of heart tissues revealed significantly reduced scar formation in animals treated with chemical cocktail (Fig. 1f, g; Supplementary information, Figure S8d). Cardiac functions were also evaluated with echocardiography. Similar extent of left ventricular dysfunctions as measured by ejection fraction (EF) and fractional shortening (FS) was observed in both groups before treatment, indicating similar level of injury following MI. Drug treatment significantly improved the heart function (Supplementary information, Figure S8e, f).
In this report, we present the first evidence of chemical-induced direct conversion of adult cardiac fibroblasts into cardiomyocytes in vivo. An interesting difference between our method and the transcription factor-based methods is that our chemical cocktail can induce cardiac reprogramming in normal mice. The previously reported in vivo cardiac reprogramming all required injury-induced fibroblast activation to allow viral infection.4, 5 The in vivo chemical-induced cardiac reprogramming is surprisingly fast, the conversion could be observed as early as 1 week after drug treatment, and prolonged treatment yielded CiCMs highly similar to adjacent cardiomyocytes. This transdifferentiation process possibly benefits from the native cardiac environment, including the extracellular matrix, secreted factors, mechanical signals, and cell–cell communications.12
CRFVPTM treatment significantly reduced the scar formation and improved cardiac functions in MI mice, although the reprogramming efficiency is only 1%. The CiCMs may only contribute partially to reduced scar and improved function. Several compounds that we used to induce transdifferentiation, including VPA, Forskolin and Rolipram, have been reported to protect cardiomyocytes and prevent collagen formation.13–15 The combination of the CiCMs, reduced fibrotic environment and protection of remaining cardiomyocytes eventually leads to a better recovery in the CRFVPTM-treated MI mice. Hopefully in near future, with improved reprogramming efficiency, the contribution of these CiCMs in cardiac function can be precisely evaluated.
In conclusion, our study is a proof of concept that chemical cocktails could be used to induce somatic cell reprogramming in vivo and our chemical combination provides a starting point for further optimization to induce cardiac regeneration.
Electronic supplementary material
Supplemental Information, Figure and Data
Acknowledgements
This work was supported by the Ministry of Science and Technology of China (2015CB964503, 2017YFA0104002), the Chinese Academy of Sciences (XDA16010202, XDA16010201), and the National Natural Science Foundation of China (81425024, 81472862, and 31501189).
Competing interests
The authors declare no competing interests.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0036-4.
References
- 1.Giudicessi JR, Kullo IJ, Ackerman MJ. Mayo Clin. Proc. 2017;92:642–662. doi: 10.1016/j.mayocp.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xin M, Olson EN, Bassel-Duby R. Nat. Rev. Mol. Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixit P, Katare R. Stem Cell Res. Ther. 2015;6:26. doi: 10.1186/s13287-015-0010-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jayawardena TM, et al. Circ. Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian L, et al. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfram JA, Donahue JK. J. Am. Heart Assoc. 2013;2:e000119. doi: 10.1161/JAHA.113.000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie X, Fu Y, Liu J. Curr. Opin. Genet. Dev. 2017;46:104–113. doi: 10.1016/j.gde.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Fu Y, et al. Cell Res. 2015;25:1013–1024. doi: 10.1038/cr.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao N, et al. Science. 2016;352:1216–1220. doi: 10.1126/science.aaf1502. [DOI] [PubMed] [Google Scholar]
- 10.Kong P, et al. Am. J. Physiol. Heart C. 2013;305:H1363–H1372. doi: 10.1152/ajpheart.00395.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang FL, Fang M, Yutzey KE. Nat. Commun. 2017;8:712. doi: 10.1038/s41467-017-00840-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riem Vis PW, et al. Eur. J. CardioThorac. 2011;39:8–17. doi: 10.1016/j.ejcts.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 13.Braunig JH, Albrecht-Kupper B, Seifert R. Naunyn Schmiede. Arch. Pharmacol. 2014;387:389–398. doi: 10.1007/s00210-013-0943-3. [DOI] [PubMed] [Google Scholar]
- 14.Kwak HJ, et al. Cell Signal. 2008;20:803–814. doi: 10.1016/j.cellsig.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Lee TM, Lin MS, Chang NC. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H968–H977. doi: 10.1152/ajpheart.00891.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information, Figure and Data