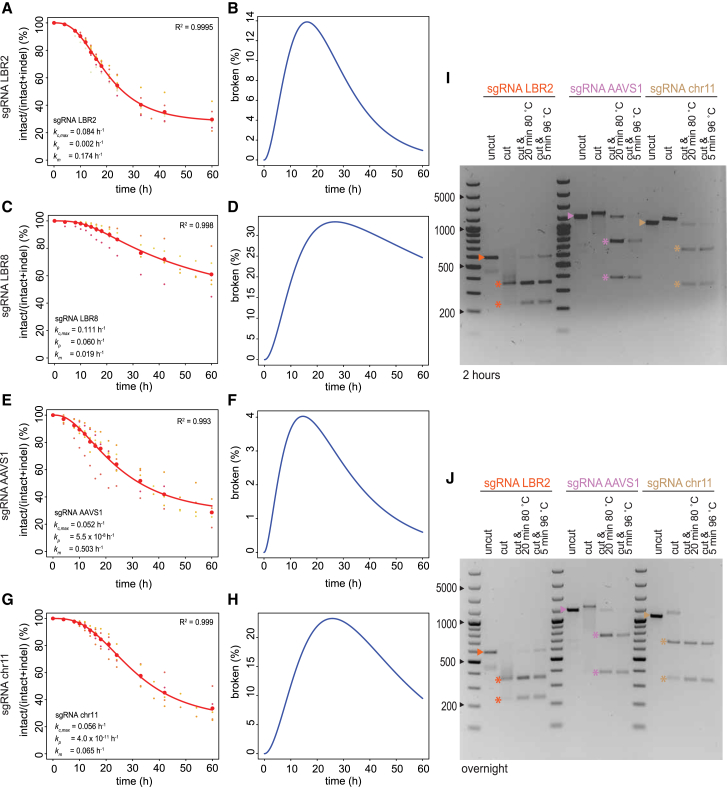
Figure 6.
ODE Modeling of DSBs at Additional Loci; Cas9 Remains Bound to Broken Ends
(A–H) Time series of Cas9 cutting and repair at four loci (LBR2, A and B; LBR8, C and D; AAVS1, E and F; intergenic region on chromosome 11, G and H).
(A, C, E, and G) Intact fraction abundance of all individual replicates is shown in small dots where each replicate has a unique color. The mean of each time point is shown as big red dots. Fitted models are shown as red curves.
(B, D, F, and H) Predicted broken fraction curves.
(I and J) In vitro digestion of DNA fragments by Cas9 and one of the sgRNAs targeting LBR (orange), AAVS1 (purple), or intergenic region on chromosome 11 (brown), respectively. DNA was incubated for either 2 hr (I) or overnight (J) in the absence (uncut) or presence (cut) of Cas9 and sgRNA (Cas9•sgRNA). In some samples, Cas9 was subsequently denatured by two different heat treatment protocols as indicated. The expected band for the intact DNA is marked by an arrowhead, and the expected digestion products are marked by asterisks.