Figure 1.
Truncated crRNAs Mediate Gene Disruption Comparable with the Full-Length Natural crRNA
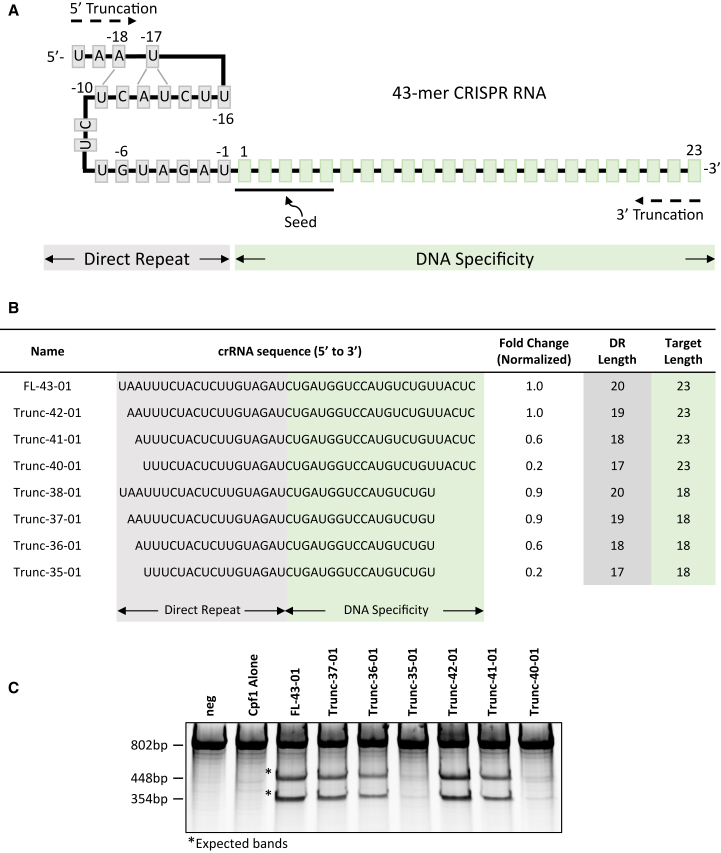
(A) Sequence recognition and structure of AsCpf1 43-mer crRNA with 5′ direct repeat indicated by nucleotides in gray box (all figures) and DNA specificity region indicated by nucleotides in light green box (all figures), including seed sequence (underlined). (B) Sequence of truncated DNMT1 targeting crRNAs where the fold change column is quantified from surveyor nuclease assay using genomic DNA isolated from transfected cells and normalized to full-length 43-mer crRNA (FL-43-01) as 1 (100%). Last two columns indicate the nucleotide length of the specified regions. Representative data from two or more replicate experiments. (C) Representative surveyor nuclease assay gel where “neg” indicates no transfection and AsCpf1 alone indicates transfection of the plasmid encoding the nuclease in the absence of crRNA. Asterisks (*) indicate expected bands.