Figure 2.
Synthetic crRNAs with Terminal 2′-O-Me Nucleotides Can Mediate Gene Disruption in Human Cells
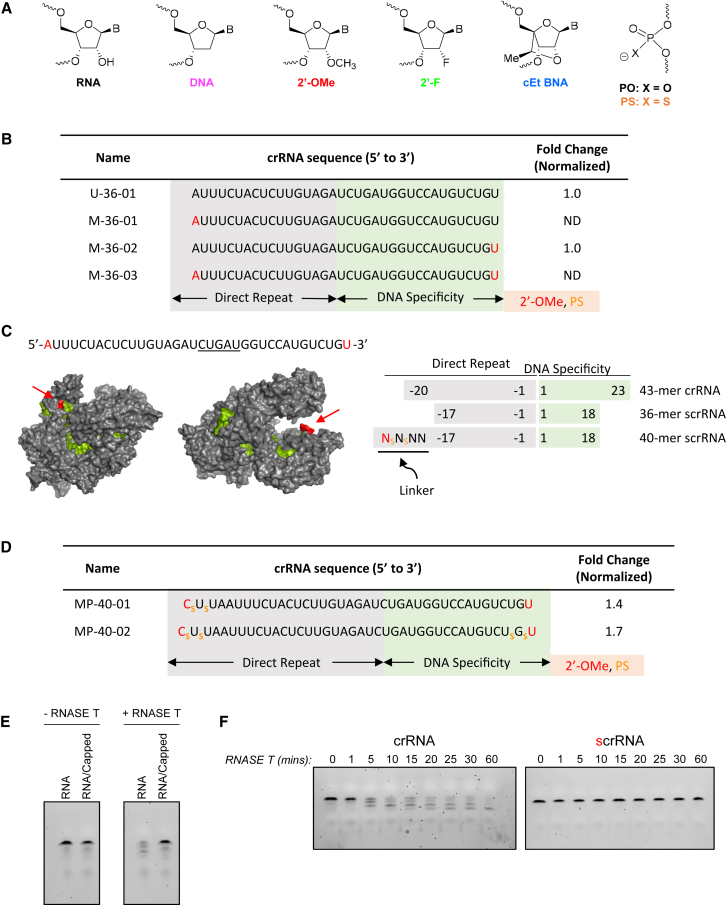
(A) Structures of chemically modified nucleotides substituted in scrRNAs including phosphorothioate (PS), 2′-O-Methyl (2′-O-Me), 2′-Fluoro (2′-F), or S-constrained ethyl (cEt) substitutions. (B) Full sequence of scrRNAs targeting DNMT1 with 2′-O-Me-substituted nucleotides indicated in red. Data representative of gene disruption quantified from surveyor nuclease assay from two or more replicate experiments and normalized to U-36-01. (C) Surface representation of AsCpf1-crRNA-target DNA complex (from PDB: 5b43)25 with the red arrow indicating the 5′ and 3′ ends of the crRNA (left). (Right) Schematic representation of scrRNAs with variable length direct repeat and DNA specificity region. 43-mer crRNA represents the natural crRNA of the AsCpf1 system. “Linker” nucleotides are underlined, and chemical substitutions are indicated by color as in (B). (D) Full sequence of DNMT1 targeting scrRNAs and single-experiment gene disruption activity compared with U-36-01 (normalized as 1). (E) SYBRgold-stained TBE-urea denaturing gel for visualization of RNA degradation products after a 30-min co-incubation with RNase T. (F) Gel same as in (E) except with the addition of multiple time points up to 60 min.