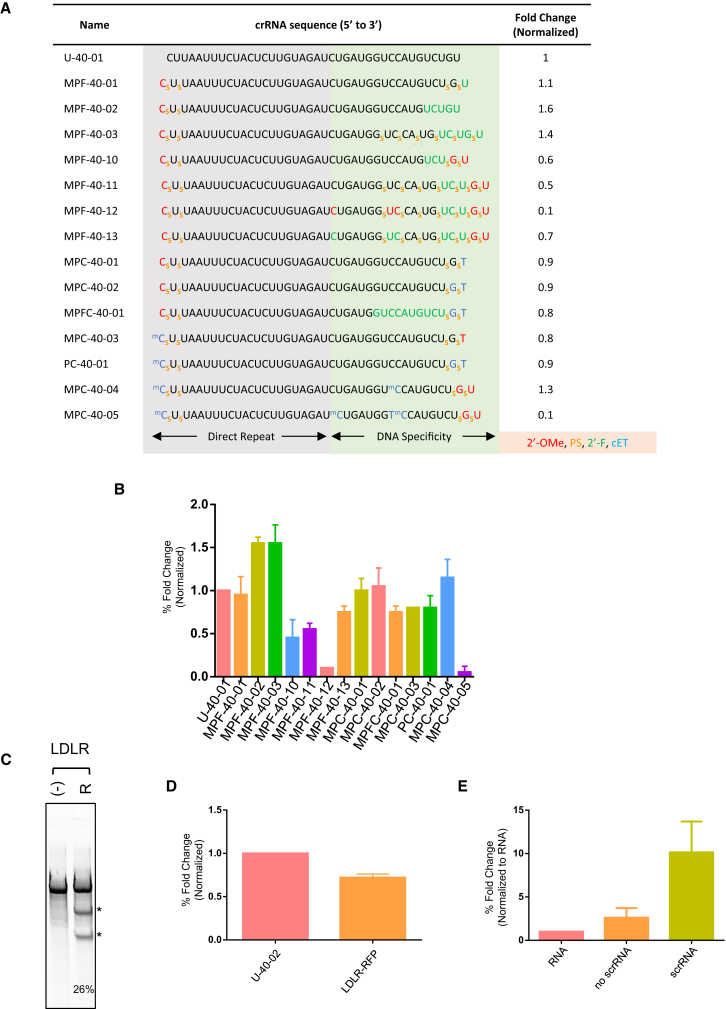
Figure 4.
Additional Chemically Modified scrRNAs Mediate Comparable Gene Disruption to Natural crRNA
(A) Complete DNMT1 target sequence with 2′-F nucleotides indicated in green, 2′-O-Me in red, and cEt in blue. Gene disruption activity normalized to U-40-01 (= 1). Data represent gene disruption quantified from surveyor nuclease assay (representative gel in Figure S6) from two or more replicate experiments. (B) Graph shows fold change in gene disruption activity as indicated in last column of (A). (C) Representative surveyor nuclease assay gel for gene editing at the LDLR locus with expected bands indicated by an asterisk (*). (D) Graph shows fold change in LDLR gene disruption activity quantified from surveyor nuclease assay from two replicate experiments. (E) Graph shows quantification of TTR RNA levels measured in triplicate from three biological replicates and normalized to neg (no transfection). No scrRNA bar represents transfection of plasmids at 0 hr as indicated in Figure S7B, but no scrRNA. scrRNA bar represents transfection of plasmids at 0 hr, and addition of scrRNA at 24 hr as indicated in Figure S7B.