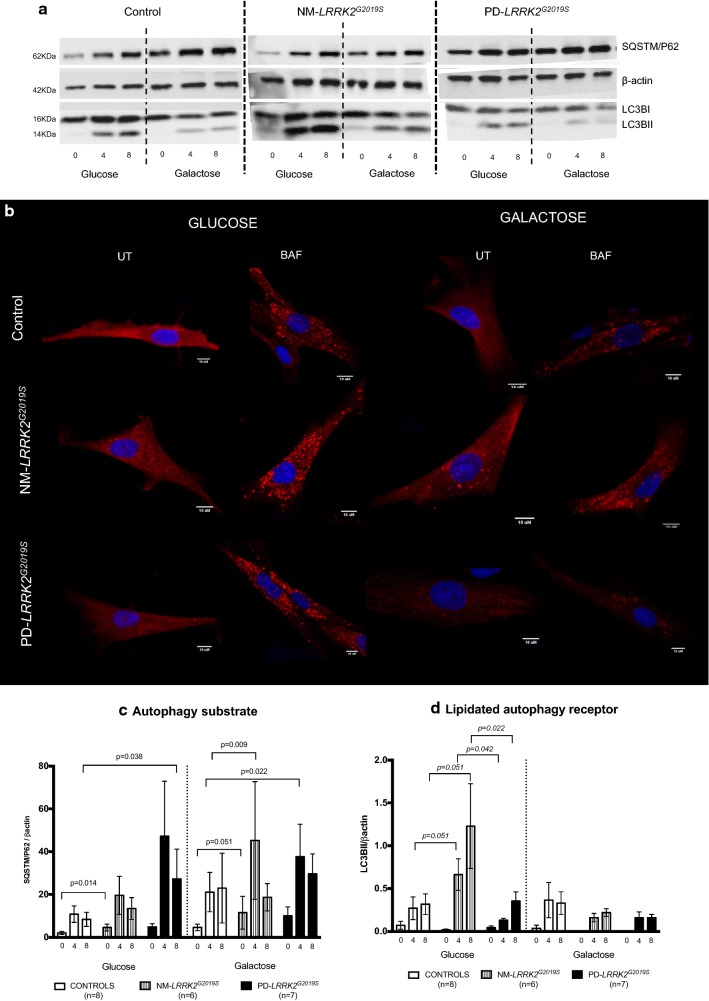
Fig. 4.
Autophagy. a Representative images (original images have been cropped and are available at request) of autophagy markers measured by Western blot comparing: NM-LRRK2G2019S: Non-manifesting carriers of LRRK2G2019S-mutation; PD-LRRK2G2019S: patients with LRRK2G2019S-mutation and clinically manifest PD. (SQSTM1/p62)/β-actin: Autophagy substrate, LC3B-I/β-actin: autophagy receptor, basal form. LC3B-II/β-actin: autophagy receptor, lipidated form. b Representative images of autophagosome formation (UT) and accumulation after 8 h of treatment with 100 nM bafilomycin (BAF) obtained by confocal microscopy. c, d Results are represented by mean ± SEM, comparing controls (n = 8; white bars), NM-LRRK2G2019S-mutation (n = 6, gray bars) and PD-LRRK2G2019S (n = 7; black bars) in either glucose or galactose media and at basal state, and after 4 and 8 h treatment with bafilomycin (0, 4, 8). Autophagy initiation was upregulated in NM-LRRK2G2019S subjects in glucose and galactose media when compared to controls (+ 118.4% SQSTM1/P62, p = 0.014 and + 114.44% SQSTM1/P62, p = 0.009, respectively) and trends to increase autophagy substrate were observed in PD-LRRK2G2019S in both media (c). Autophagosome formation trended to increase at 4 and 8 h of bafilomycin treatment in NM-LRRK2G2019S and was significantly decreased in PD-LRRK2G2019S when compared to NM-LRRK2G2019S subjects in glucose media at 4 and 8 h of bafilomycin treatment (− 79 to 86%, p = 0.042 and − 71.26%, p = 0.22 respectively) (d)